Figure 1. Growth inhibition screens in Escherichia coli reveal 218 active compounds, whose interactions with essential proteins are predicted by combining AlphaFold2 with molecular docking.
-
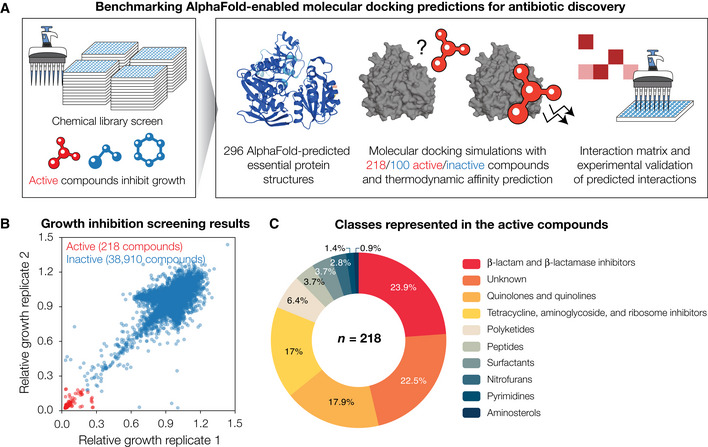
ASchematic of the approach. To define our chemical space of interest, we performed high‐throughput screens of growth inhibition against wild‐type E. coli. Compounds that inhibited growth were taken as active, and each active compound was computationally docked with each of 296 AlphaFold2‐predicted E. coli essential protein structures. For comparison, a subset of the inactive compounds was docked in the same way. An interaction matrix showing the thermodynamic binding affinities predicted by the docking simulations was then constructed. A protein‐ligand interaction was predicted to occur if its predicted binding affinity was smaller than a threshold value. All possible interactions for a subset of essential proteins, including those not predicted to occur, were empirically tested to benchmark model performance.
-
BGrowth inhibition measurements for 39,128 compounds, from which 218 compounds (including known antibiotics) were identified as active against E. coli BW25113. Data are shown from two biological replicates. Compounds with mean relative growth less than 0.2 were classified as active (red points), and all other compounds were classified as inactive (blue points).
-
CDistribution of the compound classes represented in the 218 active compounds.
