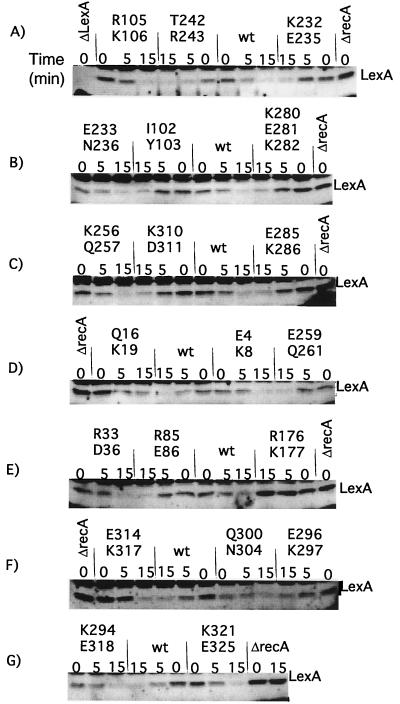
FIG. 5.
In vivo cleavage of LexA by mutant RecA proteins. Derivatives of strain JAM444 carrying pTrecA220 or mutant derivatives were grown and treated as described in Materials and Methods; the strain marked ΔrecA carried pBR322. Each panel represents the results of a representative experiment. For each mutant strain, a time course is shown. The lanes marked “0” contain samples taken without treatment; those marked “5” and “15” were taken 5 and 15 min after the addition of nalidixic acid (50 μg/ml). Samples were analyzed by Western blotting with antibody to LexA, as described in Materials and Methods. The name of the mutation is shown above each set of samples. The location of intact LexA is shown; the other bands are proteins that cross-reacted with the LexA antibody. Cleavage fragments are not shown; their levels are uninformative, since LexA cleavage fragments are unstable (27) (for an example, cf. Fig. 6). As described in the text, several mutants had less basal cleavage than the wild type. The amount of basal LexA cleavage did not appear to depend on the amount of RecA mutant protein. Q16K19 had very low levels of basal cleavage yet had higher levels of mutant RecA. On the other hand, K294E318 protein was present at very low levels and yet had the same amount of basal cleavage of LexA as the wild type (wt). The fact that the amount of basal cleavage did not correlate with the amount of RecA suggests that the extent of basal cleavage was limited by the amount of signal in the cell and/or by the ability of RecA protein to become activated by it.