Figure 1. Sup-tRNAs suppressed the mIdua-W401X mutation in HEK293 cells.
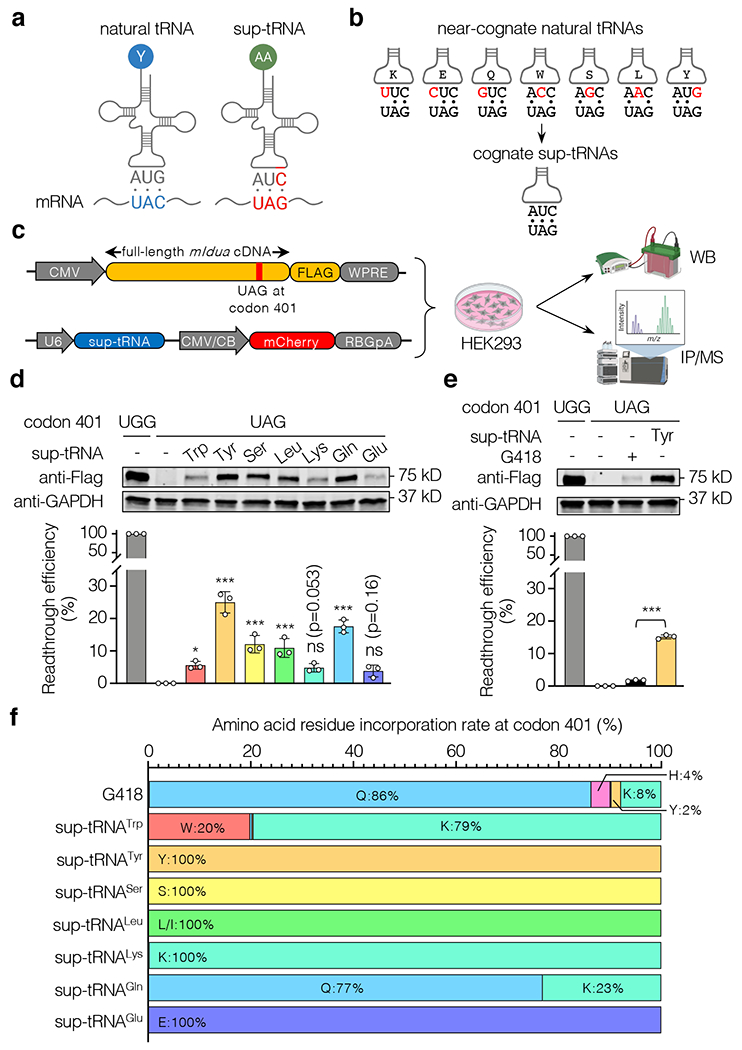
a, Schematic showing that a natural tRNA recognizes a sense codon (UAC in blue), whereas a suppressor tRNA (sup-tRNA) recognizes a stop codon (UAG in red) by the engineered anticodon. b, Near-cognate natural tRNAs have anticodons partially base-pairing with UAG (mismatches are in red), whereas sup-tRNAs have the engineered 3’-AUC-5’ anticodon that completely base-pairs with UAG. c, Workflow to examine sup-tRNA-induced readthrough efficiency by Western blot (WB) and amino acid residue incorporation by immunoprecipitation/mass spectrometry (IP/MS). Constructs expressing FLAG-tagged mouse IDUA and sup-tRNA, respectively, are shown. d, Representative Western blot images and quantification of mouse IDUA protein expression restored by sup-tRNA-induced readthrough. Data are mean ± s.d. of three biological replicates. e, Representative Western blot images and quantification of mouse IDUA protein expression restored via readthrough by G418 (0.1 mg/mL) or sup-tRNATyr. Data are mean ± s.d. of three biological replicates. f, Amino acid residue incorporation rate at the mIdua-W401X PTC by G418 or various sup-tRNAs as determined by mass spectrometry analysis of FLAG-tagged mouse IDUA. Statistical analysis was performed by one-way ANOVA followed by two-sided Dunnett’s multiple comparisons test (d and e). *p < 0.05, **p < 0.01, ***p < 0.001, ns: not significant. For gel source data, see Supplementary Figure 1.
