Extended Data Figure 1. Sup-tRNAs suppressed UAG PTCs in HEK293 cells.
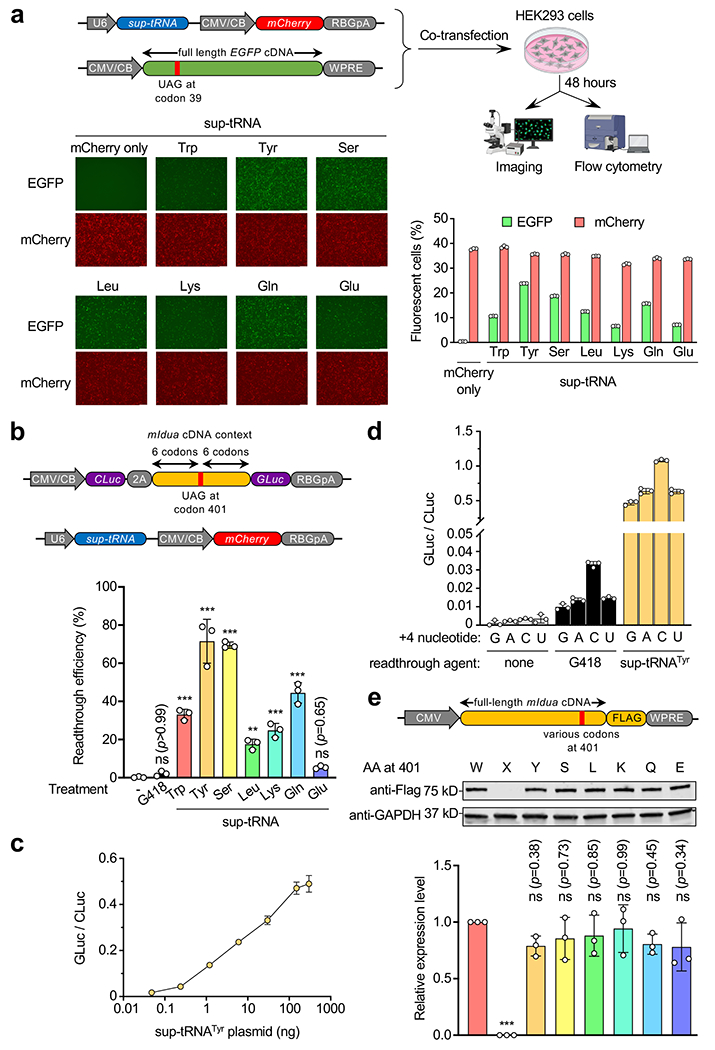
a, Workflow of a fluorescence reporter assay to examine sup-tRNA-induced readthrough of the Y39X premature termination codon (PTC) in the enhanced green fluorescence protein (EGFP) gene. Representative fluorescence images and quantification of fluorescent cells by flow cytometry are shown. Scale bar=250 μm. b, Dual-luciferase reporter assay to quantify G418 (0.1 mg/mL) or sup-tRNA-induced readthrough at the Idua-W401X PTC. Readthrough efficiency is calculated as the normalized ratio of Gaussia luciferase (Gluc) activity to Cypridina luciferase (Cluc) activity. Data are mean ± s.d. of three biological replicates. c, Dual-luciferase reporter assay using escalating amounts of sup-tRNATyr plasmid in co-transfection. d, Dual-luciferase reporter assay using a series of constructs harboring different nucleotides immediately downstream of the W401X PTC (+4 nucleotide), in the absence or presence of G418 (0.1 mg/mL) or sup-tRNATyr plasmid co-transfection. e, Representative Western blot images and quantification of protein expression from mouse Idua cDNA variants that encode different amino acid residues at codon 401. Data are mean ± s.d. of three biological replicates. Statistical analysis was performed by one-way ANOVA followed by two-sided Dunnett’s multiple comparisons test (b and e). **p < 0.01, ***p < 0.001, ns: not significant. Nucleotide sequences of EGFP-Y39X and dual-luciferase reporters are shown in Supplementary Table 4. For gel source data, see Supplementary Figure 1.
