Fig. 2.
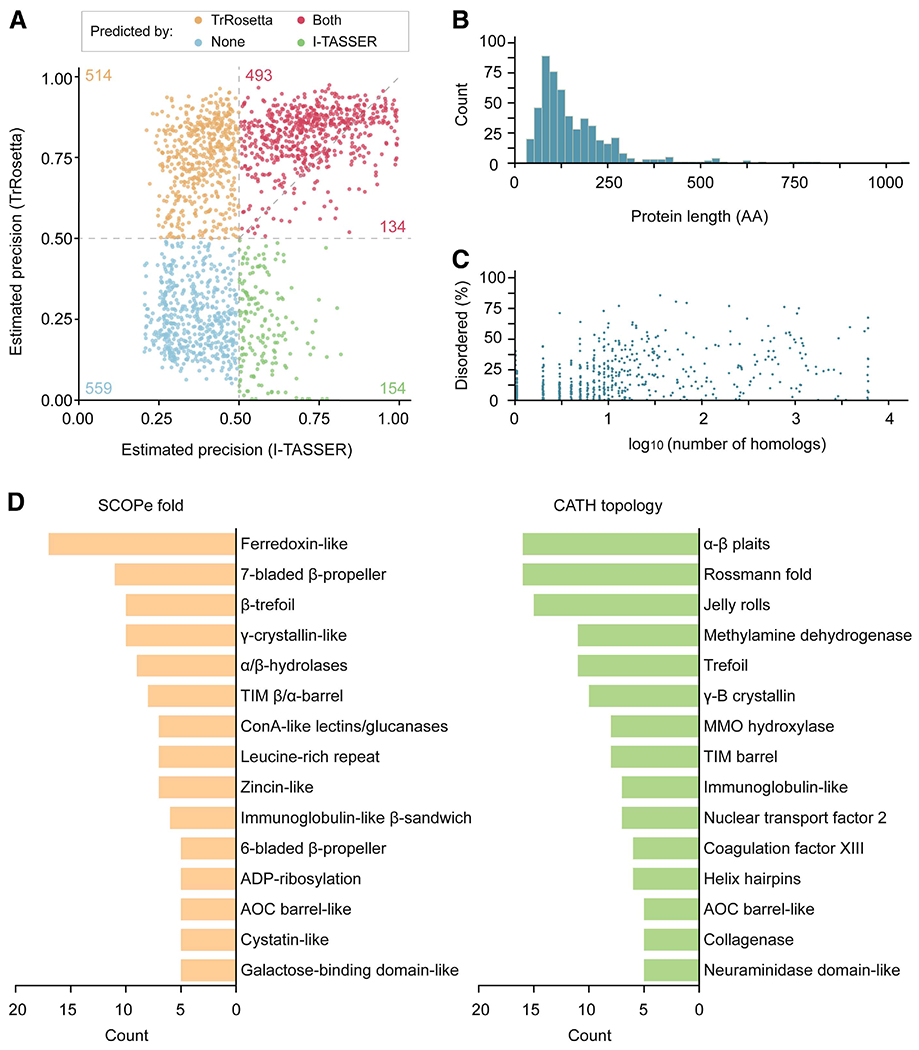
Structure prediction statistics on 1,854 Magnaporthe oryzae secreted proteins. A, Structure prediction statistics on 1,854 M. oryzae secreted proteins. The average probability of the top L predicted long- plus medium-range contacts (|i – j| >12) and the mean estimated template modeling (TM) score were used as the expected precision for TrRosetta and I-TASSER, respectively. These values are reported to well correlate with the actual precision (Yang et al. 2020; Zhang 2008). The number of proteins belonging to each section is indicated in the plot. B, Mature protein length distribution (AA = amino acids) and C, number of homologs collected for coevolutionary inference and the proportion of predicted disordered residues for 559 proteins that were not predicted by both TrRosetta and I-TASSER. D, Structure-based classification of 527 proteins without functional annotations. The 527 protein structures were assigned to SCOPe and CATH categories with RUPEE. The TM score cut-off >0.5 was required for the classification. The top 15 hits are reported.
