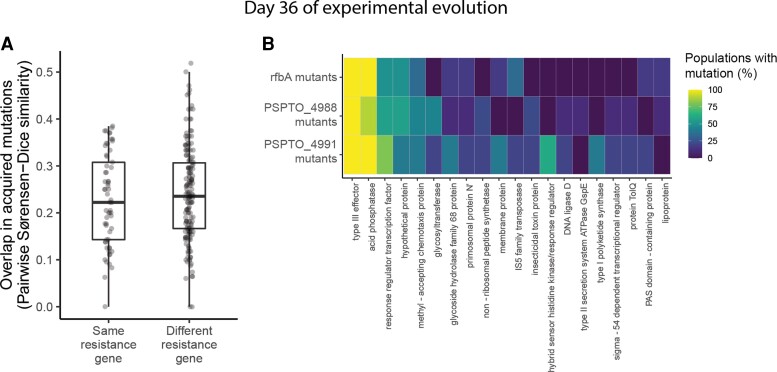
Fig. 5.
Genetic contingency is no longer detectable after extended evolution in the absence of phages. (A) Pairwise similarity coefficients among pairs of evolved populations at day 36 of experimental evolution. Statistical significance was assessed by randomizing whether pairs were labeled as having the same or different resistance genes and recalculating their similarity coefficients for 10,000 permutations. (B) Heatmap depicting the relationship between initial resistance genes and mutations acquired during experimental evolution for rfbA mutants (n = 6), PSPTO_4988 mutants (n = 9), and PSPTO_4991 mutants (n = 5). Rows represent all populations with resistance mutations in the same gene (note that resistance genes represented by fewer than two populations are not pictured, as there was no way to assess parallelism in these cases). Columns represent the top 20 genes that were most frequently mutated across populations by day 36 of experimental evolution. Note that these are not necessarily identical to the top 20 genes identified in figure 4. Colors indicate the percentage of populations of each resistance gene that had acquired one or more mutations by day 36 of experimental evolution.