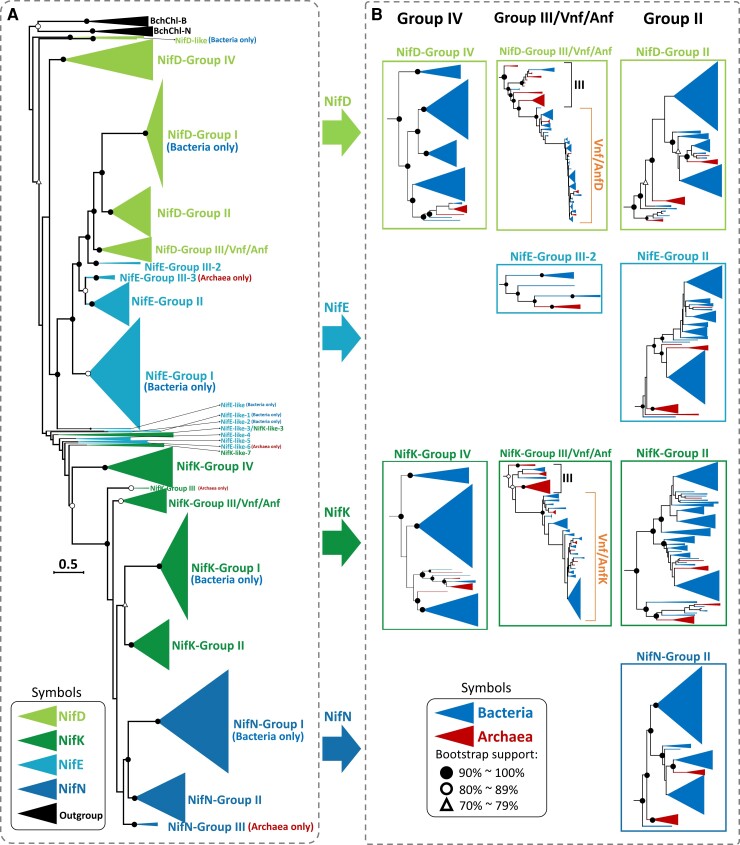
Fig. 4.
Phylogeny of 7,421 Nif/Vnf/AnfD, K, E, and N protein sequences. The scale bar denotes 0.5 amino acid substitutions per residue site. The solid and empty black circles denote the bootstrap values of 90–100% (solid circle) and 80–89% (empty circle) and the empty triangle denotes a bootstrap value of 70–79%. The light-independent protochlorophyllide reductase subunit N (Bch/ChlN) and light-independent protochlorophyllide reductase subunit B (Bch/ChlB) are used as the outgroup. The protein sequences are classified into Groups I, II, III, IV, Vnf, and Anf. (A) The outgroup is labeled by black color, NifD by light green, NifK by dark green, NifE by light blue, and NifN by dark blue. (B) is an expanded version of each subtree contained both bacteria and archaea in (A). The bacterial group is labeled by blue color and archaeal groups by dark red color. We use the IQ-TREE method to reconstruct the phylogenetic tree and ModelFinder to select the best-fit model of protein sequence evolution with ultrafast bootstrap (1,000 replicates). The model selected is LG + F + R10.