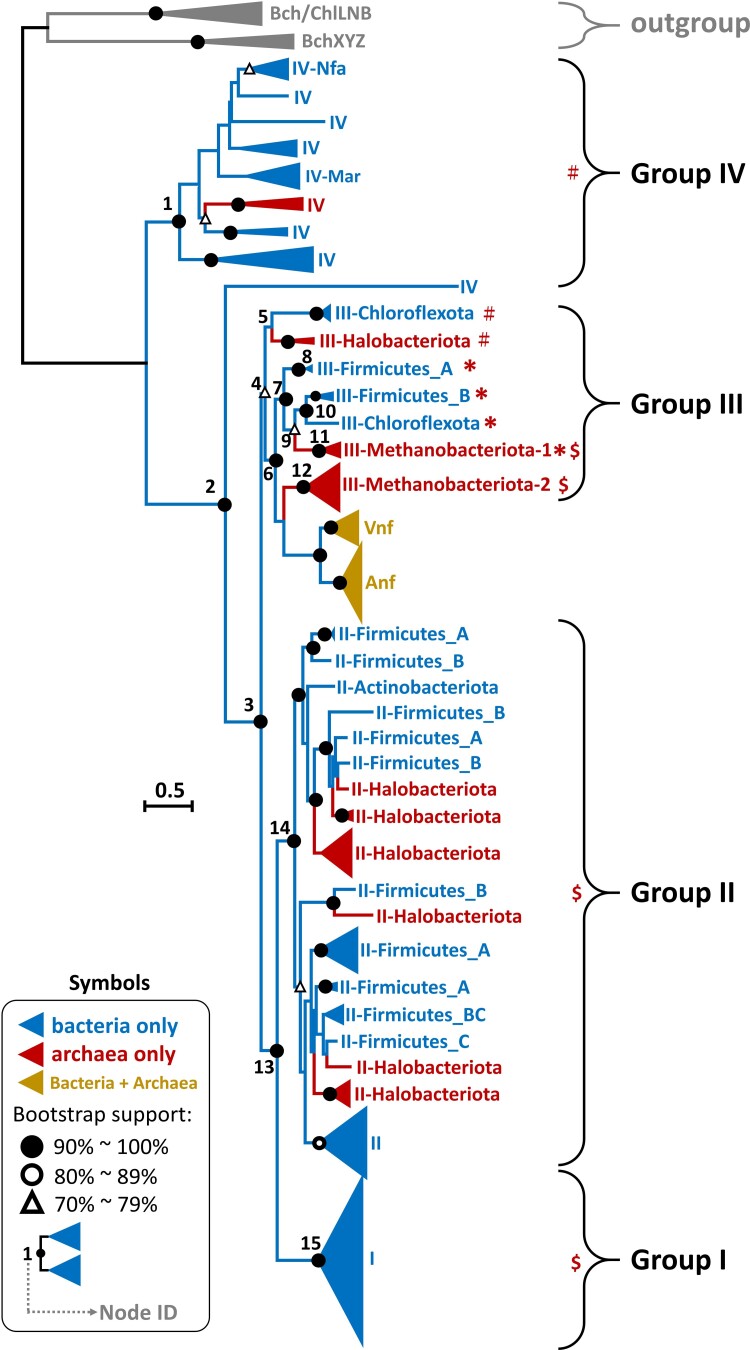
Fig. 7.
The phylogeny of 326 concatenated Nif/Vnf/AnfHDK sequences. The protein sequences are classified into Groups I, II, III, IV, Vnf, and Anf. We use the full-version ML method to construct the tree and ModelFinder to select the best-fit model of protein sequence evolution with bootstrap (500 replicates). The model selected is LG + G + I. The scale bar denotes 0.5 amino acid substitutions per residue site. The solid and empty black circles denote the bootstrap values of 90–100% and 80–89%, respectively, and the empty triangle denotes a bootstrap value of 70–79%. The bacteria are labeled by blue color and the archaea by dark red color. The Vnf/Anf groups contain both bacteria and archaea, labeled by earthy yellow. The four-nif-gene set is labeled by #, the five-nif-gene set by *, and the six-nif-gene set by $. We use 6 concatenated BchXYZ sequences and 10 concatenated Bch/Chl LNB sequences as the outgroup.