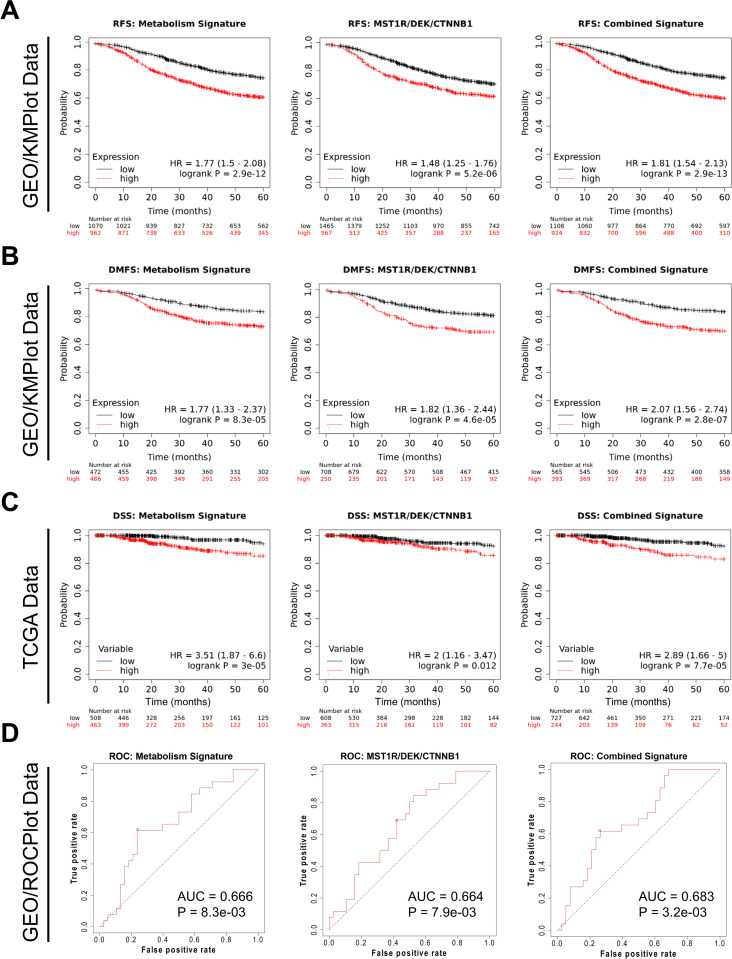
Fig 5. A metabolic gene signature developed from epistatic genes nominated by the RON-DEK-β-catenin metabolomics data predicts breast cancer patient outcomes.
Gene Expression Omnibus-derived KMPlot breast cancer datasets [22] were used to query (A) Relapse-free survival (RFS) of breast cancer patients stratified by expression of the Metabolism signature (left; see S2–S6 Figs, Table 1), RON-DEK-β-catenin (MST1R/DEK/CTNNB1) signature (middle), and both combined (Combined Signature, right) and (B) Distant metastasis-free survival (DMFS) of breast cancer patients stratified by expression of the Metabolism signature (left), RON-DEK-β-catenin (MST1R/DEK/CTNNB1) signature (middle), and both combined (Combined Signature, right). Gene signatures developed on the KMplot cancer datasets were also tested against breast cancer dataset from the Cancer Genome Atlas (TCGA) Pan Cancer datasets to examine (C) Disease-specific survival (DSS) of breast cancer patients stratified by expression of the Metabolism signature (left), RON-DEK-β-catenin (MST1R/DEK/CTNNB1) signature (middle), and both combined (Combined Signature, right). (D) Receiver-operator characteristic (ROC) analysis of node-positive breast cancer patient response to chemotherapy stratified by expression of the Metabolism signature (left), RON-DEK-β-catenin (MST1R/DEK/CTNNB1) signature (middle), and both combined (Combined Signature, right).