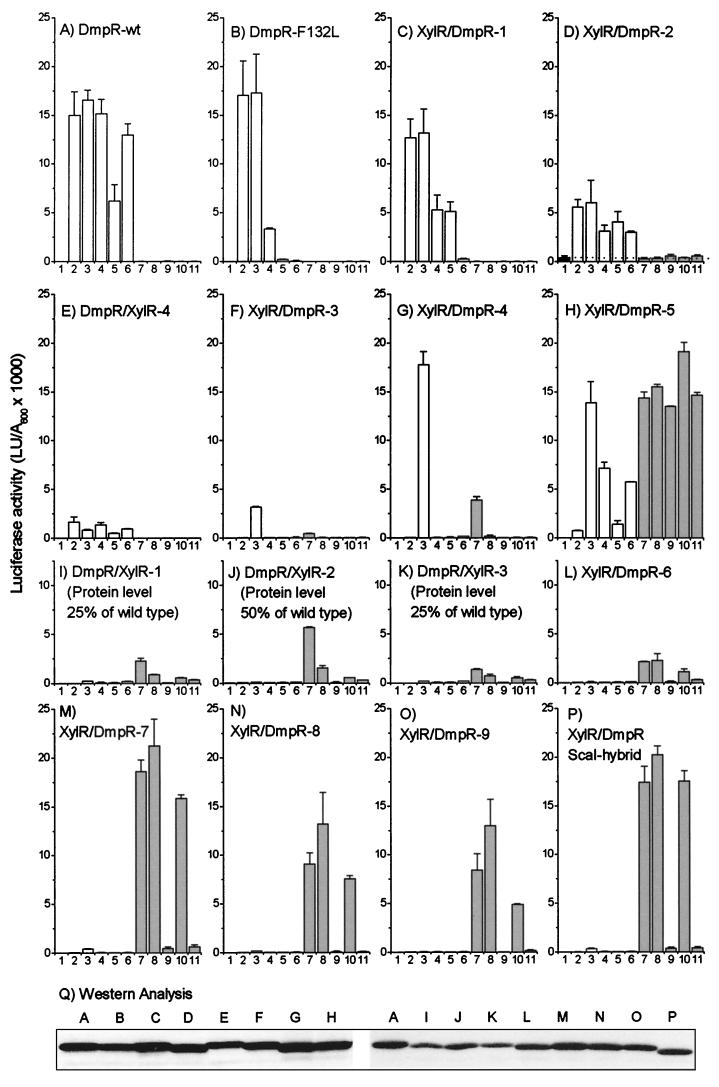
FIG. 3.
In vivo transcriptional response of Po mediated by DmpR and XylR hybrids. (A to P) Luciferase transcriptional response of P. putida KT2440::Po-luxAB harboring plasmids expressing the indicated derivatives (for plasmid designations, see Table 1) was measured in the absence of any effector (bars 1) or in the presence of two sets of aromatic effectors. The DmpR effectors (open bars) were phenol (bars 2), 2-methylphenol (bars 3), 3-methylphenol (bars 4), 4-methylphenol (bars 5), and 3,4-dimethylphenol (bars 6). The XylR and XylR-like effectors (shaded bars) were toluene (bars 7), m-xylene (bars 8), 2-methylbenzyl alcohol (bars 9), 3-methylbenzyl alcohol (bars 10), and 4-methylbenzyl alcohol (bars 11). The data are the averages of triplicate determinations in each of two independent experiments. The level of protein expression, estimated from panel Q (as described in Materials and Methods), is expressed as a percentage of that of DmpR-wt (wild type) for derivatives with significantly decreased expression levels (I, J, and K). (Q) Expression levels of the regulators. Western analysis of 30 μg of crude extract derived from cells exposed to 2-methylphenol shown in panels A to P, separated by SDS–11% PAGE, and probed with anti-DmpR serum as described in Materials and Methods is shown. LU, luciferase units. The error bars indicate standard deviations.