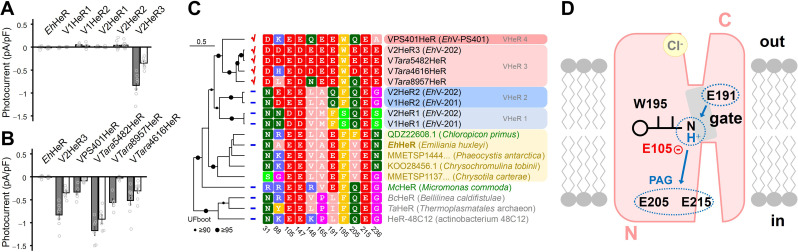
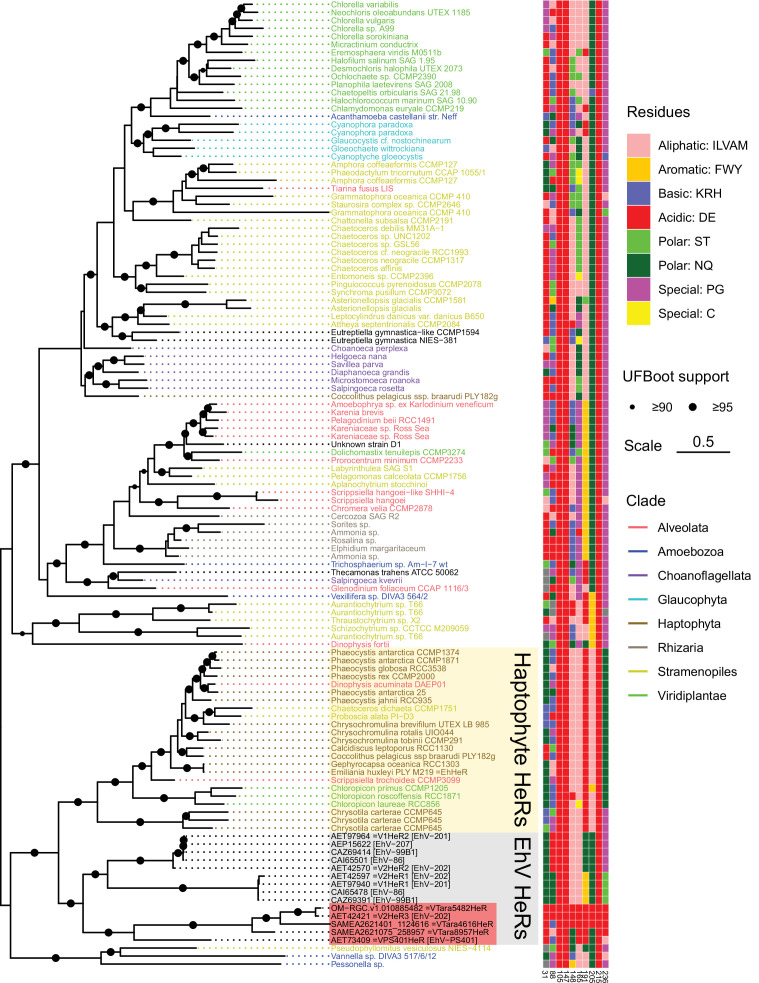
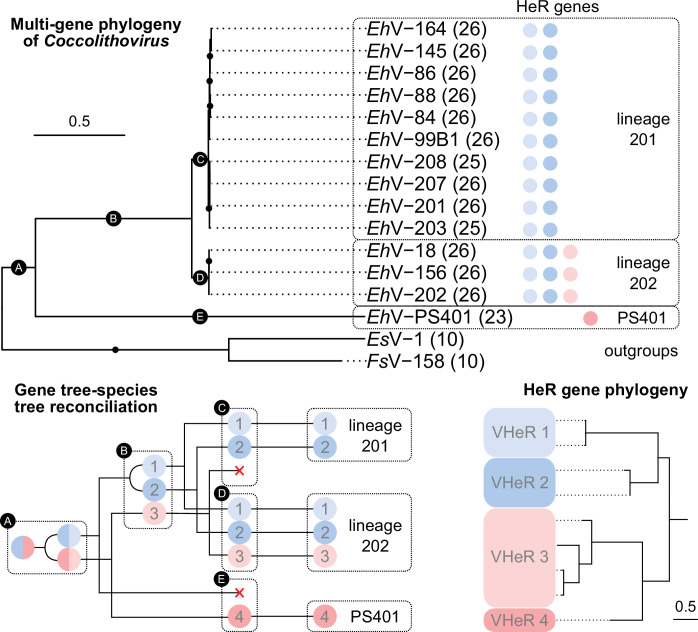
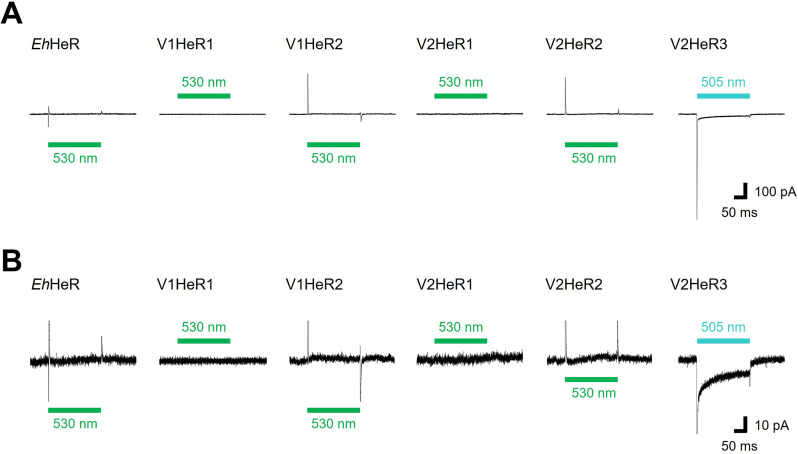
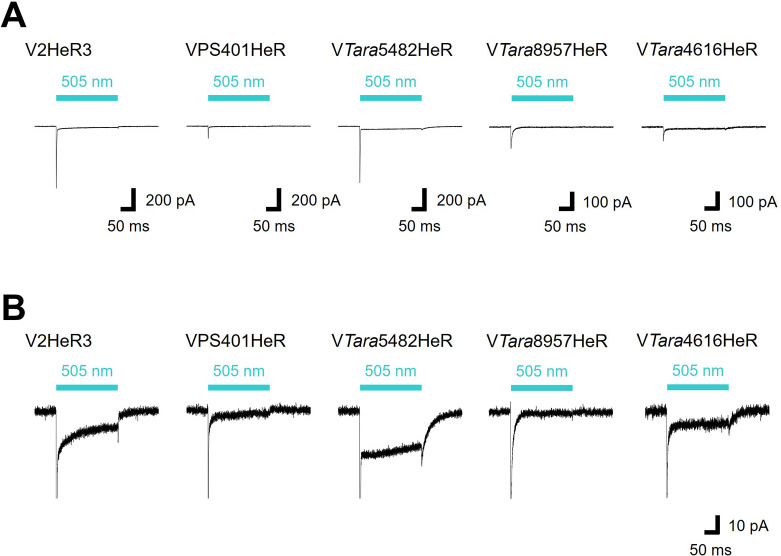
Figure 4. Electrophysiological measurements of related proteins of viral HeR from Emiliania huxleyi virus 202 (V2HeR3) and a functional model.
(A) Electrophysiological measurements of a heliorhodopsin (HeR) from the host Emiliania huxleyi (Ehux), two HeRs from E. huxleyi virus 201 (V1HeR1, 2) and three HeRs from E. huxleyi virus 202 (V2HeR1-3). Although some HeRs exhibit transient photocurrents (positive or negative peaks), steady-state photocurrent was only observed for V2HeR3. Comparison of photocurrent densities at −40mV. Square-block bar indicates amplitude from peak photocurrent (I1), open bar indicates amplitude from steady-state photocurrent (I2). The pipette solution was 110mM NaCli, pH 7.4i, the bath solution was 140mM NaCle, pH 7.4e. n=5–8cells. (B) Electrophysiological measurements and the obtained I-V plots of homologous proteins of V2HeR3. Comparison of photocurrent amplitudes at −40mV. Square-block bar indicates amplitude from peak photocurrent (I1), open bar indicates amplitude from steady-state photocurrent (I2). The pipette solution was 110mM NaCli, pH 7.4i, the bath solution was 140mM NaCle, pH 7.4e. n=5–8cells. (C) Key residues for ion transport of HeRs. (D) Schematic drawing of suggested proton-transporting mechanism in V2HeR3.