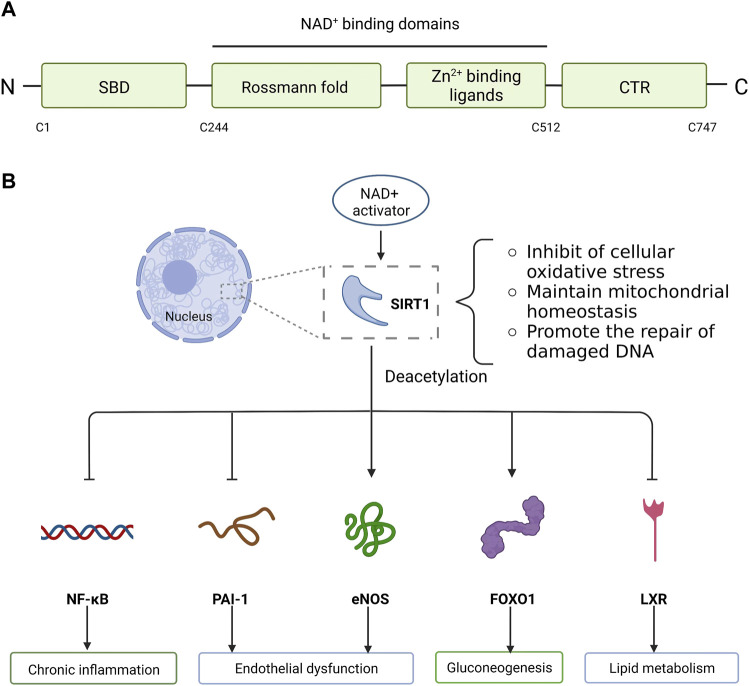
FIGURE 1.
Molecular structure and biological function of SIRT1. (A) SIRT1 is composed of N-terminal, C-terminal and NAD+ dependent catalytic core region. The catalytic core region (C244-C512) is folded into two subdomains: Zn2+ binding ligands and Rossmann fold conformation. N-terminal contains SBD and C-terminal contains CTR. (B) SIRT1, mainly located in the nucleus, deacetylates related proteins and reduces cell apoptosis by inhibiting cellular oxidative stress, maintaining mitochondrial metabolic homeostasis, and promoting the repair of damaged DNA. SIRT1 activity is dependent on NAD+. The activation of SIRT1 is facilitated by increasing NAD+ levels at the cellular level, which can lead to deacetylation and modulated expression of many downstream targets. SIRT1 targets a variety of substrates and performs different functions. SIRT1 deacetylates inflammation-related transcription factor NF-κB, which attenuates NF-κB driven inflammation. In addition, SIRT1 protects endothelial cells against replicative senescence by deacetylating eNOS and downregulating PAI-1 expression. In the liver, SIRT1 deacetylates and activates the transcription factor FOXO1 to stimulate gluconeogenesis. Similarly, SIRT1 regulates lipid metabolism by modulating LXR via deacetylation of this molecular receptor. SIRT1, Sirtuin1; SBD, sirtuins-activating compounds binding domain; CTR, C-terminal regulatory segment; NAD+, nicotinamide adenine dinucleotide; NF-κB, nuclear factor kappa B; eNOS, endothelial nitric oxide synthase; PAI-1, plasminogen activator inhibitor 1; FOXO1, forkhead box O 1; LXR, liver X-receptor.