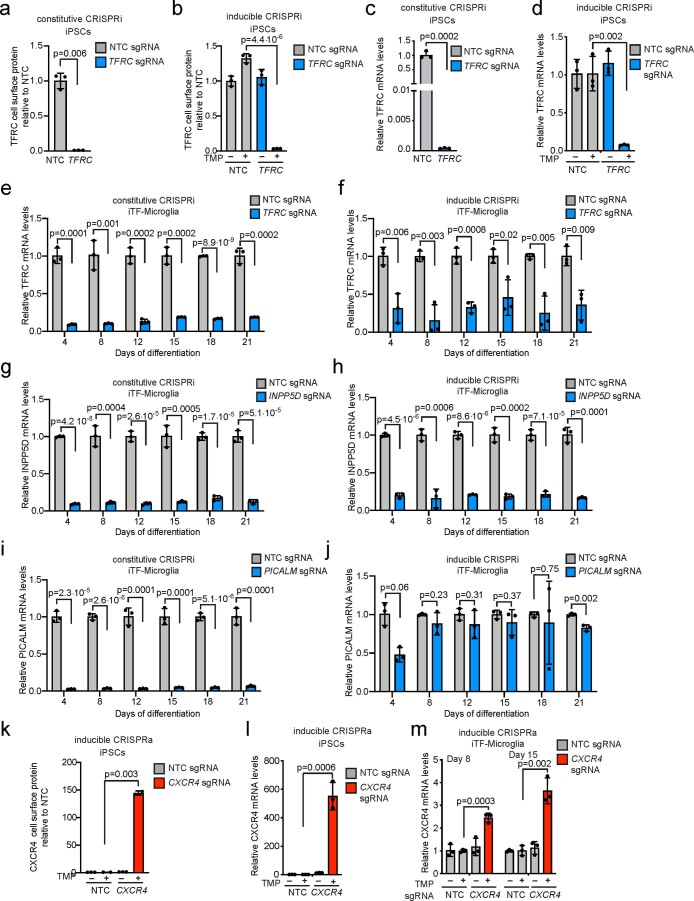
Extended Data Fig. 4. Functional validation of CRISPRi/a activity in iPSCs and iTF-Microglia.
a, b, Functional validation of constitutive (a) or inducible (b) CRISPRi activity via flow cytometry of TFRC surface protein level stained iPSCs expressing a TFRC-targeting sgRNA or a non-targeting control (NTC) sgRNA (mean + /− sd, n = 3 biological replicates; p values from two-tailed Student’s t-test). TMP was added to induce CRISPRi activity where indicated. c-d, Knockdown of TFRC in iPSCs with (a) the constitutive and (b) the inducible CRISPRi system. qPCR quantification of the relative fold change of TFRC mRNA levels in CRISPRi-iPSCs expressing a TFRC sgRNA as compared to a non-targeting control sgRNA in the presence or absence of trimethoprim (TMP). (mean + /− sd, n = 3 biological replicates; p values from two-tailed Student’s t-test). TFRC levels were normalized to the housekeeping gene GAPDH. e-j Knockdown of three different genes in iTF-Microglia with (e,g,i) constitutive CRISPRi and (f,h,j) inducible CRISPRi. qPCR quantification of the relative fold change of TFRC mRNA levels (e,f), INPP5D mRNA levels (g,h) or PICALM mRNA levels (I,j) in CRISPRi-iTF-Microglia expressing a TFRC sgRNA (e,f), INPP5D sgRNA (g,h) or PICALM sgRNA (I,j) compared to a non- targeting control sgRNA at different days of differentiation in the presence of TMP (mean + /− sd, n = 3 biological replicates, P values from two-sided Student’s t test). k, Functional validation of inducible CRISPRa activity via flow cytometry of CXCR4 surface protein level stained iPSCs expressing a CXCR4-targeting sgRNA or a non-targeting control (NTC) sgRNA (mean + /− sd, n = 3 biological replicates; p values from two-tailed Student’s t-test). TMP was added to induce CRISPRi activity where indicated. l-m, qPCR quantification of the relative fold change of CXCR4 mRNA levels in inducible CRISPRa-iPSCs expressing a CXCR4 sgRNA as compared to a non-targeting control sgRNA in the presence or absence of trimethoprim (TMP), which stabilizes the DHFR degron. (mean + /− sd, n = 3 biological replicates; p values from two-tailed Student’s t-test). CXCR4 levels were normalized to the housekeeping gene GAPDH.