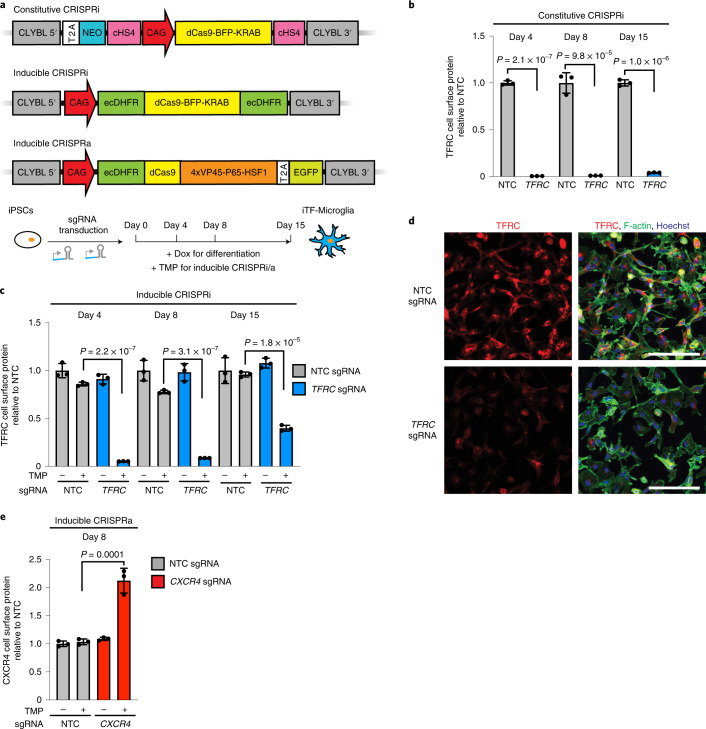
Fig. 3. Gene knockdown and overexpression by CRISPRi and CRISPRa in iTF-Microglia.
a, Strategies for constitutive and inducible CRISPRi/CRISPRa in iTF-Microglia. Top, for constitutive CRISPRi, a dCas9-BFP-KRAB construct (catalytically dead Cas9 (dCas9) fused to BFP and the KRAB transcriptional repressor domain) is expressed from the constitutive CAG promotor integrated into the CLYBL safe-harbor locus. Middle, for inducible CRISPRi, dCas9-BFP-KRAB is tagged with ecDHFR degrons. Bottom, for inducible CRISPRa, CAG promotor-driven ecDHFR-dCas9-VPH was stably integrated into the CLYBL locus. VPH, activator domains containing 4× repeats of VP48, P65 and HSF1. Addition of TMP stabilizes the inducible CRISPRi/a machineries. b,c, Functional validation of constitutive (b) or inducible (c) CRISPRi activity via flow cytometry of TFRC surface protein level stained iTF-Microglia expressing a TFRC-targeting sgRNA or an NTC sgRNA at different days of differentiation (mean ± s.d., n = 3 biological replicates; P values from two-tailed Student’s t-test). c, TMP was added to induce CRISPRi activity where indicated. d, Functional validation of inducible CRISPRi activity via TFRC immunofluorescence (IF) microscopy on day 8. Top row, NTC sgRNA. Bottom row, sgRNA targeting TFRC. TFRC, red; F-actin, green; nuclei, blue. Scale bar, 100 μm. e, Functional validation of inducible CRISPRa activity via flow cytometry of CXCR4 surface protein level staining in iTF-Microglia expressing CXCR4 sgRNA or NTC sgRNA (mean ± s.d., n = 3 biological replicates; P values from two-tailed Student’s t-test). TMP was added to induce CRISPRa activity where indicated.