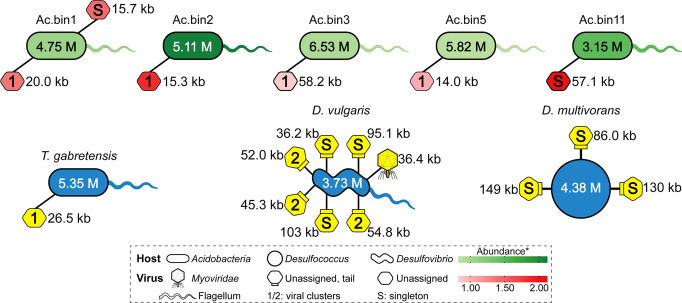
Fig. 3. Predicted linkages between SRMs and their viruses.
The 16 medium- to high-quality MAGs of SRMs were included in the analysis. For comparison, the complete genomes of T. gabretensis, D. vulgaris and D. multivorans were chosen. The cell shapes of the SRMs were drawn based on information from the literature and from our analysis (e.g., the presence or absence of core genes encoding the flagellum). The predicted genome sizes of SRMs and their viruses are shown. Increasing abundances of the SRMs are indicated by darker green colors. The abundance was calculated as the normalized mean coverage depth. Whenever applicable, viral shapes were drawn based on taxonomic information from the signature genes detected in the viral scaffolds. Otherwise, viruses are indicated by hexagons. The hexagons associated with a small rectangle represent tailed viruses. The number (i.e., 1 or 2) within a given virus represents a viral cluster, from which the virus was derived. That is, those hexagons filled with the same number are affiliated with the same viral cluster. Singletons represent new viruses. Increasing abundances of the acidobacterial viruses are indicated by darker red colors. Due to the lack of abundance information, the three cultured species and their viruses are shown in blue and yellow, respectively. Additional details are presented in Supplementary Table 9.