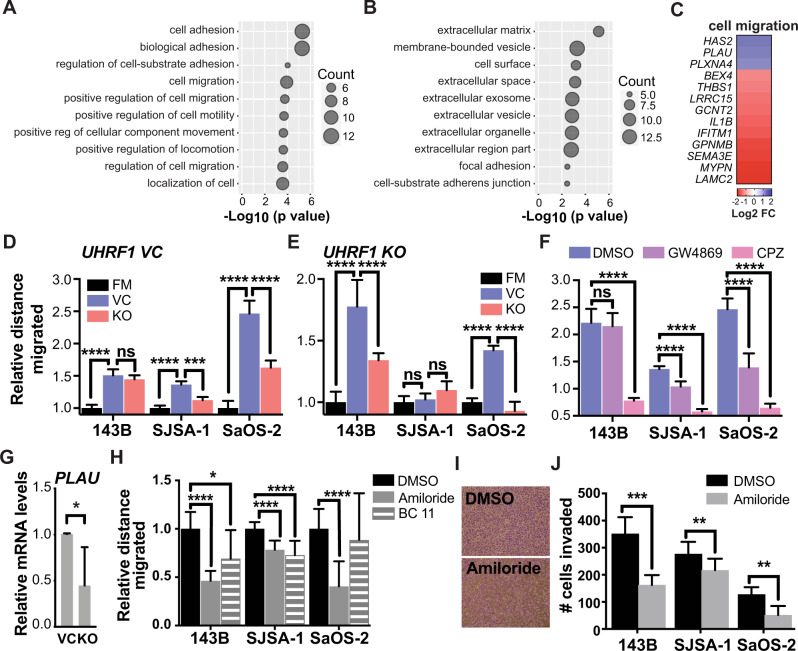
Fig. 4. UHRF1 controls the expression of genes involved in exosomes and urokinase plasminogen activator production to drive osteosarcoma migration.
A, B Gene ontology (GO) analysis for (A) biological processes and (B) cellular components of the downregulated differentially expressed genes (DEGs) identified through RNA-seq. C Heatmap of top 13 genes involved in the regulation of cell migration, decreased (blue) or increased (red) in expression level upon UHRF1 loss. D, F Quantification of relative distance migrated in scratch-wound assays for (D) osteosarcoma VC cells assayed with fresh media (FM) or conditioned media (24 h) from VC cells or UHRF1 KO cells, normalized to FM; E osteosarcoma UHRF1 KO cells assayed with FM or conditioned media from VC cells or UHRF1 KO cells, normalized to FM; F each of the osteosarcoma cell lines treated with DMSO, 10 µM GW4869 or 20 µM CPZ, normalized to DMSO control. Each data point is mean ± s.d. of ten measurements in triplicate samples. G qPCR analysis of PLAU mRNA levels in flank-implanted tumors from control (VC) and UHRF1 KO (KO) SJSA-1 cell lines. *P < 0.05 by paired two-tailed t test. H Quantification of relative distance migrated in scratch-wound assays for each of the osteosarcoma cell lines treated with DMSO, 150 µM amiloride, or 16.4 µM BC11 hydrobromide, normalized to DMSO control. Each data point is mean ± s.d. of ten measurements in triplicate samples. I Representative images from Transwell assays with cells stained with crystal violet comparing levels of invasion between cells treated with DMSO or 150 µM amiloride in SJSA-1. J Quantification of the number of cells invaded in Transwell invasion assay for each of the osteosarcoma cell lines treated with DMSO or 150 µM amiloride. Each data point is mean ± s.d. of triplicate samples. For all graphs: ns not significant, **P < 0.01, ***P < 0.001, ****P < 0.0001 by unpaired two-tailed t test.