Abstract
Uterine leiomyomas, or fibroids, are the most common tumors in women of reproductive age. Uterine leiomyomas can be classified into at least three main molecular subtypes according to mutations affecting MED12, HMGA2, or FH. FH-deficient leiomyomas are characterized by activation of the NRF2 pathway, including upregulation of the NRF2 target gene AKR1B10. Here, we have identified a novel leiomyoma subtype showing AKR1B10 expression but no alterations in FH or other known driver genes. Whole-exome and whole-genome sequencing revealed biallelic mutations in key genes involved in neddylation of the Cullin 3-RING E3 ligase, including UBE2M, NEDD8, CUL3, and NAE1. 3′RNA sequencing confirmed a distinct molecular subtype with activation of the NRF2 pathway. Most tumors displayed cellular histopathology, perivascular hypercellularity, and characteristics typically seen in FH-deficient leiomyomas. These results suggest a novel leiomyoma subtype that is characterized by distinct morphological features, genetic alterations disrupting neddylation of the Cullin 3-RING E3 ligase, and oncogenic NRF2 activation. They also present defective neddylation as a novel mechanism leading to aberrant NRF2 signaling. Molecular characterization of uterine leiomyomas provides novel opportunities for targeted treatment options.
Subject terms: Cancer genomics, Gynaecological cancer
Introduction
Uterine leiomyomas, or fibroids, are benign tumors that arise from the smooth muscle wall of the uterus. They are the most common neoplasms affecting women during reproductive years. Although non-cancerous, leiomyomas frequently cause a variety of symptoms including pressure upon adjacent organs, abnormal uterine bleeding, pelvic pain, and impaired fertility [1]. Leiomyomas are the leading indication for hysterectomy worldwide and they pose a significant socio-economic burden [2]. Approximately 10% of leiomyomas can be distinguished from “conventional” leiomyomas, as they display variant histopathology or distinct growth patterns [3]. Common histopathological variants include cellular leiomyomas, leiomyomas with bizarre nuclei, and mitotically active leiomyomas. Although these display some histopathological features associated with malignancy, they are considered clinically benign [3].
Recent studies have revealed the existence of various molecular leiomyoma subtypes. Indeed, 80–90% of leiomyomas harbor one of three genetic changes: a hotspot mutation in mediator complex subunit 12 (MED12), a chromosomal aberration resulting in upregulation of high mobility group AT-hook 2 (HMGA2), or biallelic loss of fumarate hydratase (FH) [4]. Pathogenic FH mutations may also be inherited, causing Hereditary Leiomyomatosis and Renal Cell Cancer (HLRCC) [5]. This syndrome is characterized by multiple uterine and cutaneous leiomyomas, as well as renal cell cancer of papillary type 2 histopathology. Mutations in genes encoding for members of the SRCAP histone-loading complex were recently discovered as a rare fourth molecular subtype [6]. While the known leiomyoma driver mutations have been discovered in most conventional tumors, a substantially larger proportion of leiomyoma variants lack them [7]. Cellular leiomyomas have been associated with 1p loss, but no well-established driver gene has been identified [8].
Uterine leiomyoma subtypes have been shown to display distinct global gene expression patterns [6, 9]. Leiomyomas of the FH subtype are characterized by activation of nuclear factor, erythroid 2 like 2 (NRF2) target genes, including upregulation of aldo-keto reductase family 1 member B10 (AKR1B10) and NAD(P)H quinone dehydrogenase 1 (NQO1) [9, 10]. NRF2 is an antioxidant defense transcription factor that is regulated by a Cullin-RING ligase (CRL) E3 ligase complex containing kelch like ECH associated protein 1 (KEAP1) as a substrate-recognition module and cullin 3 (CUL3) as a scaffold protein [11]. Excessive levels of fumarate result in protein succination, which is a chemical modification that occurs when fumarate reacts with cysteine residues to generate S-(2-succinyl)cysteine (2SC). Succination of critical cysteine residues within KEAP1 impairs its ability to direct NRF2 for ubiquitination and degradation by the proteasome, resulting in stabilization of de novo synthesized NRF2 [12, 13]. Oncogenic activation of the NRF2 pathway is a common feature in many cancer types [14]. While cancers frequently harbor loss-of-function mutations in KEAP1 and CUL3 or gain-of-function mutations in NFE2L2 (encoding NRF2) itself [15], no such mutations have been detected in uterine leiomyomas. We and others have shown that overexpression of AKR1B10 can function as a biomarker for FH-deficiency and NRF2 activation in uterine leiomyomas and renal cell cancer [9]. Indeed, we recently analyzed publicly available leiomyoma data and reclassified two leiomyomas as FH-deficient partly based on the expression of AKR1B10 [16]. Using FH-deficient tumors derived from HLRCC patients and sporadic leiomyomas with biallelic loss of FH, we recently showed that AKR1B10 immunohistochemistry can act as a biomarker on protein-level as well [17]. In this study, we present a novel uterine leiomyoma subtype that is FH-proficient but displays AKR1B10 protein expression. We have integrated next-generation sequencing with gene expression profiling and discovered candidate driver mutations for these tumors. We have also evaluated the feasibility of AKR1B10 expression in detecting NRF2 activated tumors.
Results
Immunohistochemistry reveals AKR1B10 expression in FH-proficient leiomyomas
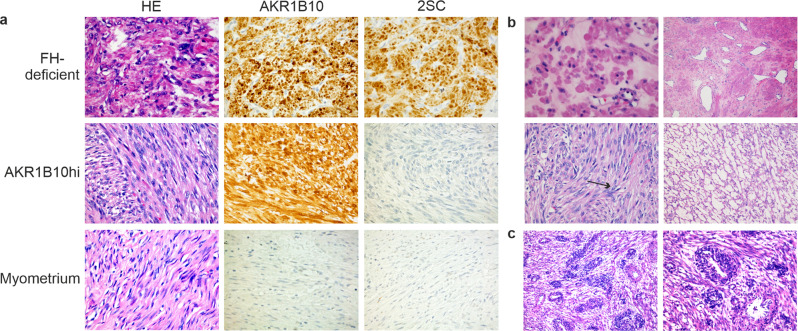
To evaluate the expression of AKR1B10 in histopathological leiomyoma variants, we screened 141 FFPE variant tumors by immunohistochemistry. This sample set has been previously examined for defects in MED12 by direct sequencing and alterations affecting HMGA2 or FH by immunohistochemistry [7, 18, 19]. As expected, all twelve FH-deficient leiomyomas displayed AKR1B10 expression (Supplementary Table 1 and Fig. 1a). In addition, we identified AKR1B10 expression in nine leiomyomas with no indication of FH-deficiency or alterations affecting MED12 or HMGA2 (Table 1 and Fig. 1a). To study this further, we analyzed another previously examined dataset of 360 unselected leiomyomas [20] and discovered three additional tumors with AKR1B10 expression and no known driver defects. These twelve tumors are hereafter referred to as AKR1B10hi leiomyomas.
Fig. 1. Immunohistochemistry and histopathological evaluation reveal AKR1B10 expression and specific morphological features in FH-proficient leiomyomas.
a 2SC and AKR1B10 antibodies can both act as biomarkers for FH-deficiency. Immunohistochemistry showed strong staining with both antibodies in FH-deficient tumors. It also revealed a subgroup of tumors positive only for AKR1B10 (AKR1B10hi). Histopathological evaluation of the AKR1B10hi tumors showed increased cellularity in most of them. Myometrium was negative for both AKR1B10 and 2SC. b Half of the AKR1B10hi tumors showed two or more features associated with FH-deficiency [37], including eosinophilic inclusions (top left), staghorn vasculature (top right), eosinophilic nucleoli and nuclear atypia (bottom left), and alveolar edema (bottom right). Example of eosinophilic nucleoli is marked with an arrow. c Half of the AKR1B10hi samples displayed perivascular hypercellularity, a feature not associated with leiomyomas previously. Magnification 100× (left) and 200× (right).
Table 1.
Immunohistochemistry of 498 uterine leiomyomas reveal AKR1B10 expression in 12 FH-proficient tumors (AKR1B10hi).
| Histopathology | N | AKR1B10hi | % | p-value** |
|---|---|---|---|---|
| Conventional | 348 | 2 | 0.6 | |
| Variant | 150 | 10 | 6.7 | 0.0002 |
| Cellulara | 97 | 10 | 10.3 | 0.00001 |
| Bizarre nuclei | 32 | 0 | 0 | 1 |
| Mitotically active | 19 | 0 | 0 | 1 |
| Epitheloid | 1 | 0 | 0 | 1 |
| Lipoleiomyoma | 1 | 0 | 0 | 1 |
aCellular leiomyomas included 12 tumors with also mitotic activity and two tumors with also bizarre nuclei. Two tumors with mitotic activity and two tumors with bizarre nuclei were AKR1B10hi tumors.
**p-values have been calculated with Fisher’s exact test. Frequency of AKR1B10hi in variant tumors has been compared to tumors with conventional histopathology.
AKR1B10hi leiomyomas display distinct morphological features
Three uterine leiomyomas were included in both previously mentioned datasets and the total sample series hence consisted of 498 tumors, 348 with conventional and 150 with variant histopathology (Supplementary Table 1). Of the twelve AKR1B10hi samples, two had been diagnosed as conventional leiomyomas and ten displayed variant histopathology as defined by WHO classification [3] (Table 1). Interestingly, all ten variant tumors displayed increased cellularity. Six of the tumors showed cellularity as the only main histopathological feature, while two tumors displayed mitotic activity and two other tumors displayed bizarre nuclei as well. Overall, AKR1B10hi tumors were thus significantly over-represented in leiomyoma variants (p-value = 0.0002, two-sided Fisher’s exact test) and even more significantly over-represented in cellular leiomyomas (p-value = 0.00001, two-sided Fisher’s exact test) compared to conventional tumors. Thorough re-evaluation of the twelve AKR1B10hi tumors revealed that six of the tumors displayed at least two histopathological characteristics typically associated with FH-deficiency (Supplementary Table 2 and Fig. 1b). In addition, six tumors showed perivascular hypercellularity, a feature not usually found in uterine leiomyomas (Fig. 1c).
Genes involved in neddylation of the Cullin 3-RING E3 ligase are recurrently mutated in AKR1B10hi leiomyomas
To identify candidate driver mutations and somatic copy number alterations (SCNA) in AKR1B10hi samples, we performed whole-exome and/or whole-genome sequencing with the twelve AKR1B10hi tumors and eight corresponding normal tissue samples. One exome tumor-normal pair was only analyzed for SCNAs as it displayed low coverage and poor sequencing quality. As expected, we detected no driver changes in FH, MED12, or HMGA2. We identified five genes that were mutated in more than one sample (Supplementary Table 3). Interestingly, two of the recurrently mutated genes encode for proteins involved in neddylation of the Cullin 3-RING E3 ligase; we identified three samples with a mutation in ubiquitin conjugating enzyme E2 M (UBE2M) and two samples with a mutation in NEDD8 ubiquitin like modifier (NEDD8) (Table 2 and Supplementary Fig. 1). Sanger sequencing revealed one additional UBE2M mutation in the tumor with poor exome-sequencing quality. These findings prompted us to search for other mutations related to neddylation among the non-recurrently mutated genes. We identified one sample with a mutation in the NEDD8-binding domain of Cullin 3 (CUL3). We then searched for additional mutations in an in-house exome sequencing dataset of seven fresh frozen leiomyomas. This revealed two mutations in NEDD8 activating enzyme E1 subunit 1 (NAE1) in one leiomyoma, which was negative for the known leiomyoma driver alterations. Histopathological evaluation of the corresponding FFPE sample revealed cellular histopathology and perivascular hypercellularity. In addition, immunohistochemistry showed strong staining for AKR1B10 adding this sample to the AKR1B10hi group. None of the identified neddylation-associated mutations were present in The Genome Aggregation Database (gnomAD). All variants were either truncating or predicted deleterious with a Combined Annotation-Dependent Depletion (CADD) value of over 20 or a Genomic Evolutionary Rate Profiling (GERP) value of over 2 [21, 22]. All mutations were validated by Sanger sequencing and confirmed as somatic where normal tissue was available (Supplementary Fig. 2).
Table 2.
Candidate driver mutations in AKR1B10hi tumors.
| Sample | Gene | Mutation | SCNA | Confirmed somatic | Prediction score |
| 1301_1_S1 | UBE2M | c.97C > T, p.(Arg33Trp) | Deletion | Yes | CADD: 28 |
| 1370_1_S1 | UBE2M | c.281A > T, p.(Lys94Met) | Deletion | Yes | CADD: 28.7 |
| 1364_1_S1 | UBE2M | c.298dupT, p.(Tyr100Leufs*6) | Deletion | Yes | NA |
| 1356_1_S1 | UBE2M | c.348_368delinsTCCG, p.(Arg116Serfs*40) | CN-LOH | Yes | NA |
| 1367_1_S1 | UBE2M | — | Deletion | — | — |
| 1285_1_S1 | NEDD8 | c.8T > C, p.(Ile3Thr) | Deletion | NA | CADD: 31 |
| 1354_1_S1 | NEDD8 | c.44T > C, p.(Ile15Thr) | Deletion | Yes | CADD: 26.8 |
| 1593_1_S1 | NEDD8 | — | Deletion | — | — |
| 1298_1_S2 | CUL3 | c.2222_2224del, p.(Ile741del) | Deletion | Yes | GERP: 5.4 |
| 1055_1_S1 | NAE1 | c.1422del, p.(Asp476Metfs*33) | — | NA | NA |
| c.740T > A, p.(Ile247Asn) | CADD: 29.4 |
SCNA somatic copy number alteration; CN-LOH copy-neutral loss of heterozygosity, NA not analyzed (germline sample not available or in silico analysis not applicable).
Three AKR1B10hi samples (1308_1_S1, 1608_1_S4, and 1626_1_S2) did not harbor any candidate driver changes related to neddylation.
A combined annotation-dependent depletion (CADD) value of over 20 indicates a deleterious change.
A genomic evolutionary rate profiling (GERP) value of over 2 indicates high evolutionary conservation.
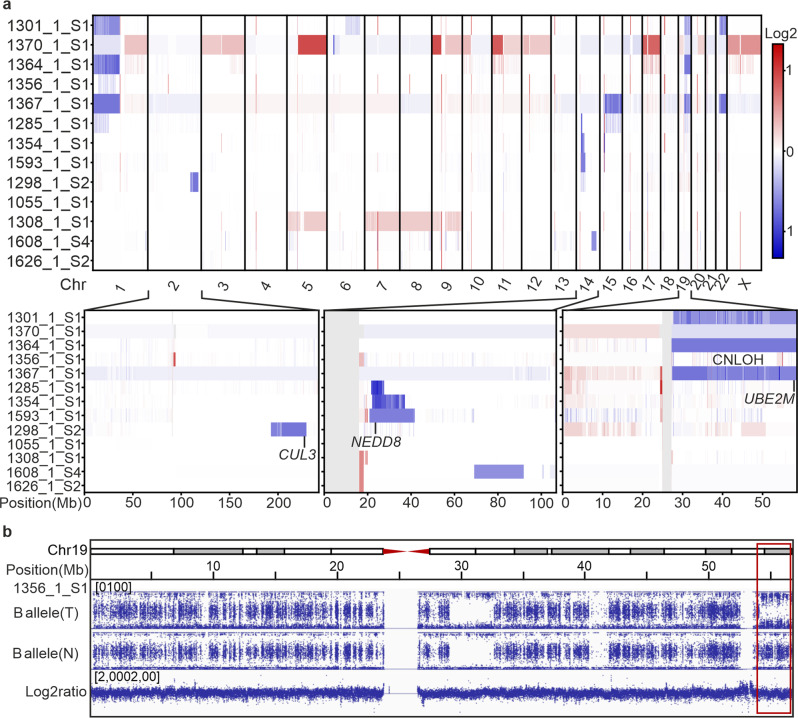
Somatic copy number alteration analysis of all thirteen AKR1B10hi tumors revealed that most chromosomes were relatively stable (Fig. 2a). However, we did identify recurrent whole-arm and terminal losses on chromosomes 1p, 19q, and 22q, as well as three interstitial deletions affecting 14q. In addition, we identified one sample with a terminal copy-neutral loss of heterozygosity (CN-LOH) event at 19q (Fig. 2b). The 19q and 14q alterations frequently overlapped with the gene-level mutations, resulting in biallelic loss of UBE2M on 19q13.43 and biallelic loss of NEDD8 on 14q12. Alterations affecting 1p, 19q, and 22q co-occurred with each other, whereas the interstitial deletions in 14q were mutually exclusive with the former changes. Only one sample displayed an interstitial deletion in 2q, resulting in biallelic loss of CUL3 (Fig. 2a). We identified no large-scale chromosomal alterations in the sample that harbored two NAE1 mutations. Large-scale amplified regions were seen in only two samples, including a shared amplification of the 5q-arm and whole-chromosome amplification of chromosome 9 (Fig. 2a).
Fig. 2. Somatic copy number alterations in AKR1B10hi leiomyomas.
a AKR1B10hi leiomyomas displayed relatively stable chromosomal profiles. Recurrent loss of the 19q-arm was seen in four AKR1B10hi leiomyomas and three of these deletions overlapped with a gene-level mutation in UBE2M. An interstitial 14q deletion was seen in three leiomyomas and two of these deletions overlapped with a gene-level mutation in NEDD8. Only one sample harbored a 2q deletion, and this deletion overlapped with a gene-level mutation in CUL3. b One sample with a gene-level mutation in UBE2M displayed copy-neutral loss of heterozygosity (CN-LOH) at the terminal q-arm of chromosome 19.
3′RNA sequencing confirms activation of the NRF2 pathway in AKR1B10hi leiomyomas
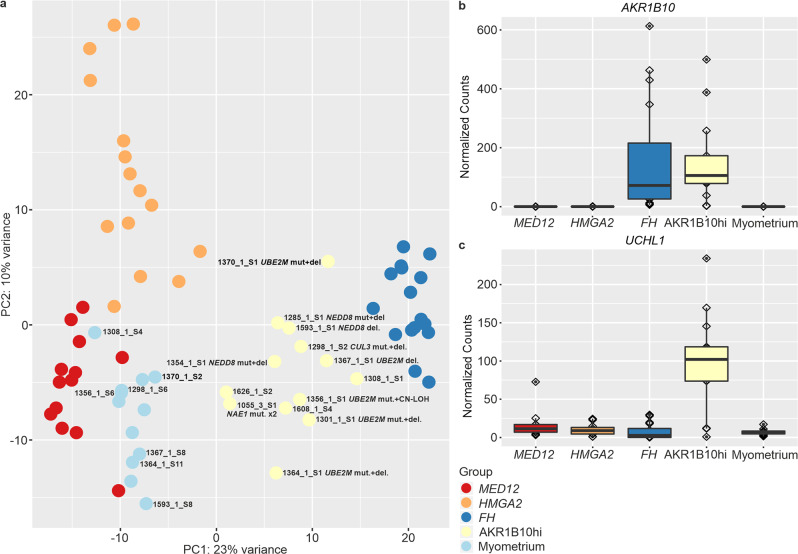
To explore the gene expression pattern of AKR1B10hi leiomyomas, we performed 3′RNA sequencing with thirteen AKR1B10hi leiomyomas and seven corresponding myometrium samples. We analyzed these samples together with a previously published dataset of 44 leiomyomas [16]. Principal component analysis revealed that the AKR1B10hi samples clustered separately from the other three established leiomyoma subtypes, but close to leiomyomas of the FH subtype (Fig. 3a).
Fig. 3. Principal component analysis and differentially expressed genes in AKR1B10hi leiomyomas.
a Thirteen AKR1B10hi leiomyomas and seven corresponding myometrium samples were analyzed together with a previously published dataset of 44 leiomyoma and 5 myometrium samples. Principal component analysis revealed that the AKR1B10hi samples cluster separately from leiomyomas of the MED12, HMGA2, and FH subtypes. Samples analyzed in this study are marked with sample identifiers. b AKR1B10 was significantly upregulated in AKR1B10hi and FH-deficient leiomyomas, but not in the other leiomyoma subtypes. c Deubiquitination gene UCHL1 was among the most uniquely upregulated genes in AKR1B10hi leiomyomas.
Differential expression analysis comparing AKR1B10hi samples against the myometrium controls revealed 1014 differentially expressed genes (q-value < 0.05; |FC | > 2, Supplementary Table 4), with AKR1B10 being the second most significant one (FC = 34.54, q-value = 4.68 × 10−25, Fig. 3b). Pathway enrichment analysis confirmed significant dysregulation of the NRF2 pathway in this novel leiomyoma subtype (q-value = 3.29 × 10−03; Supplementary Table 5), including upregulation of the well-established NRF2 target gene NQO1 (FC = 5.58, q-value = 3.07 × 10−12). Immunohistochemistry confirmed overexpression of NQO1 in 11 AKR1B10hi samples (Supplementary Fig. 3). Two AKR1B10hi samples failed in the analysis as no NQO1 expression was detected in either the internal control (endothelial) cells nor in the smooth muscle cells. To find uniquely expressed genes, we compared AKR1B10hi leiomyomas against the other leiomyoma subtypes and myometrium samples and identified 585 significant genes (Supplementary Table 6). Ubiquitin C-terminal hydrolase L1 (UCHL1) was the third most significant gene (FC = 6.11, q-value = 1.09 × 10−11, Supplementary Table 6 and Fig. 3c).
AKR1B10 is a robust biomarker for NRF2 activation across different malignancies
To determine the feasibility of AKR1B10 as a biomarker for NRF2 activation across different malignancies, we exploited The Cancer Genome Atlas (TCGA) and Cancer Cell Line Encyclopedia (CCLE) multilevel data. First, we utilized receiver-operating characteristic (ROC) analysis to investigate how AKR1B10 mRNA expression is associated with hotspot and truncating mutations in KEAP1 and hotspot mutations in NFE2L2 in all TCGA samples. AKR1B10 mRNA exhibited robust discrimination of NRF2 activation with an area under the ROC curve of 0.91 (Supplementary Fig. 4a). This was further validated at the protein level, as AKR1B10 protein expression correlated with the mRNA in CCLE data (r = 0.82, p < 0.001, Supplementary Fig. 4b).
NRF2 activation through mutations in neddylation-associated genes is rare in cancers
We utilized multilevel TCGA data to investigate whether somatic mutations in UBE2M, NAE1, NEDD8, or the NEDD8-binding domain of CUL3 can be found in cancers with NRF2 activation. Based on the ROC analysis, we defined tumors with an activated NRF2 by utilizing the true positive rate of over 0.8 for AKR1B10 expression. We then removed samples with a somatic mutation in NFE2L2, KEAP1, FH, or CUL3 (excluding the NEDD8-binding domain). This revealed 11 samples across six different cancer types with a nonsynonymous mutation in a neddylation-associated gene: four with a UBE2M mutation, five with a NAE1 mutation, one with a NEDD8 mutation, and one with a mutation in the NEDD8-binding domain of CUL3 (Supplementary Figs. 1 and 4c).
Inhibition of neddylation in melanoma cells results in activation of the NRF2 pathway
Using publicly available microarray data, we explored the global gene expression pattern of melanoma cells that had been treated with the NEDD8-activating enzyme inhibitor MLN4924 for up to 24 h. Gene set enrichment analysis (GSEA) revealed a significant enrichment of gene sets related to the NRF2 pathway (Supplementary Fig. 5a). Furthermore, we observed a gradual increase in AKR1B10 expression with a peak fold-change of 4.45 in cells that had been treated with MLN4294 compared to controls (Supplementary Fig. 5b). According to fold-change ranking at the 24 h time point, AKR1B10 was the 27th most upregulated gene among 19421 genes examined on the array.
Discussion
Uterine leiomyomas are benign smooth muscle tumors that can be classified into at least three molecular subtypes, reflecting mutations in either MED12, HMGA2, or FH [4]. We have previously shown that each defect results in a distinct global gene expression pattern [9]. Leiomyomas of the FH subtype are characterized by activation of the NRF2 pathway, including upregulation of the NRF2 target gene AKR1B10 [9]. In this study, we found AKR1B10 expression in a subset of leiomyomas with no FH mutations. These tumors were also negative for MED12 and HMGA2 defects, as well as for mutations in genes encoding for proteins of the SRCAP complex, another recently discovered leiomyoma subtype [6]. This suggests that these AKR1B10 overexpressing tumors represent a novel leiomyoma subtype.
Using next-generation sequencing, we identified biallelic mutations in key genes involved in neddylation of the Cullin 3-RING E3 ligase [23, 24]. Neddylation is analogous to ubiquitination and the process by which NEDD8 is conjugated to target proteins for degradation [25]. NEDD8 is a ubiquitin-like protein that activates the largest family of ubiquitin E3 ligases, the Cullin–RING ligases [26]. Neddylation is mediated by a NEDD8-activating enzyme E1 (NAE, a heterodimer consisting of NAE1 or UBA3), a NEDD8-conjugating enzyme E2 (UBE2M or UBE2F), and a NEDD8 ligase E3 (including the KEAP1/CUL3/RBX1 E3-ubiquitin ligase complex) [26]. Here, we identified mutually exclusive mutations in NEDD8, NAE1, and UBE2M. All mutations were either truncating or predicted deleterious and frequently accompanied by loss of heterozygosity, suggesting that they result in biallelic inactivation and defective neddylation. We were unable to find a second mutation in UBE2M in one tumor with a 19q loss and in NEDD8 in one tumor with an interstitial 14q deletion. However, deletions in 19q and 14q were mutually exclusive in all tumors studied, suggesting that they indeed target UBE2M and NEDD8, respectively. UBE2M is specifically involved in neddylation of Cullins 1–4 and interestingly, we also identified a 3 bp deletion in the NEDD8-binding domain of CUL3, suggesting that the identified mutations specifically disrupt neddylation of the Cullin 3-RING E3 ligase [27]. Indeed, a Cul3K712R mutant in the same domain has been shown to result in reduced binding of NEDD8 to CUL3 [28].
Gain-of-function mutations in NFE2L2, loss-of-function mutations in key members of the KEAP1/CUL3/RBX1 E3-ubiquitin ligase complex, and succination of KEAP1 have proven to activate the NRF2 pathway in many cancer types [14]. Our results suggest that defective neddylation represents a novel mechanism for oncogenic NRF2 activation (Fig. 4). Using 3′RNA sequencing and immunohistochemistry of NQO1, we confirmed a significant dysregulation of the NRF2 pathway in AKR1B10hi tumors. To our knowledge, mutations in neddylation-associated genes have not been previously identified as potential drivers of tumorigenesis. Using multilevel TCGA and CCLE data, we showed that AKR1B10 expression is a robust biomarker for NRF2 activation across cancers. By utilizing AKR1B10 as a biomarker for NRF2 activation, we found only 11 samples with NRF2 activation and a candidate mutation in UBE2M, NAE1, NEDD8, or the NEDD8-binding domain of CUL3. This suggests that defective neddylation is not a common mechanism driving NRF2 activation in cancers.
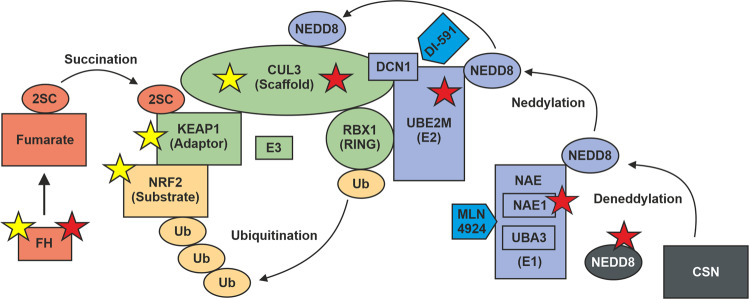
Fig. 4. Schematic illustration of mechanisms leading to oncogenic NRF2 activation.
NRF2 is, under basal conditions, ubiquitinated by the KEAP1/CUL3/RBX1 E3-ubiquitin ligase complex. Neddylation is required for the activation of cullin-RING ubiquitin ligases (CRL). The COP9 signalosome (CSN) is responsible for deneddylation of CRL. KEAP1 plays a central role in regulating the activity of NRF2. In cancers, the interaction between the E3 subunits can be disrupted by specific mutations in NFE2L2 (encoding NRF2) or by biallelic loss of KEAP1 or CUL3 (yellow stars). The interaction between NRF2 and KEAP1 can also be disrupted by protein succination of critical cysteine residues within KEAP1 as a result of fumarate accumulation. Oncogenic activation of the NRF2 pathway can be seen in uterine leiomyomas that harbor biallelic loss of FH or biallelic loss of key members involved in the neddylation pathway (red stars). Finally, inhibition of neddylation by MLN4924 or DI-591 results in activation of NRF2.
MLN4924 is a potent and selective small-molecule inhibitor of NAE1 [29]. Functional studies have shown that inhibiting neddylation by the MLN4924 inhibitor results in stabilization of the NRF2 protein [29, 30]. Using publicly available expression data of melanoma cells treated with the MLN4924 inhibitor, we confirmed significant upregulation of NRF2 target genes, including AKR1B10 as one of the most significantly upregulated genes. In addition, defective in cullin neddylation 1 (DCN1) promotes neddylation of Cullin-RING ubiquitin ligases by interacting with UBE2M [31, 32]. DCN1 is more critical in neddylation of cullin 3 than other cullin family members. DI-591 is another small-molecule inhibitor that binds to DCN1 and selectively converts cellular cullin 3 into an un-neddylated inactive form [31]. Furthermore, knockdown of DCN1 has been shown to result in accumulation of NRF2. These data support that defective neddylation leads to oncogenic NRF2 activation and strengthen the notion of neddylation-associated mutations as potential drivers in tumorigenesis.
The AKR1B10hi tumors displayed a global expression pattern distinct from the other established leiomyoma subtypes. In contrast to FH-deficient leiomyomas that also display NRF2 activation, but presumably functional neddylation, we identified the deubiquitin gene UCHL1 as one of the most uniquely upregulated genes in AKR1B10hi tumors. UCHL1 encodes for an enzyme called ubiquitin carboxyl-terminal esterase L1 (UCH-L1) that interacts with the COPS5 subunit of the COP9 signalosome (CSN) [33]. CSN is a protein complex that deactivates Cullin-RING ubiquitin ligases by catalyzing the hydrolysis of NEDD8 from the cullin subunit [34]. UCH-L1 has a high affinity for both ubiquitin and NEDD8, but it can only hydrolyze ubiquitin, unlike its more widely distributed homolog UCH-L3 [35]. The possibility of UCH-L1 exerting an effect through binding to and/or regulating NEDD8 has been proposed, but remains to be fully explored [36]. It is tempting to speculate that upregulation of UCHL1 may be a consequence of defective neddylation in these tumors.
Approximately 10% of leiomyomas display variant histopathology with increased cellularity being the most common feature. Histopathological features associated with FH-deficiency are typically seen in leiomyomas with bizarre nuclei, but rarely in leiomyomas with cellular histopathology [37, 38]. Here, we discovered more than one FH-associated feature in six AKR1B10hi tumors: in five cellular leiomyomas and in one leiomyoma with conventional histopathology. This suggests that morphological features typically seen in FH-deficient tumors may be related to activation of the NRF2 pathway itself, and not with loss of FH. The majority of AKR1B10hi leiomyomas also showed perivascular hypercellularity, a feature not typically seen in uterine leiomyomas.
Cellular leiomyomas have previously been associated with the loss of 1p, and these tumors display some gene expression patterns seen in leiomyosarcomas [39]. Multiple studies have reported 1p loss to be frequently accompanied by 19q loss in leiomyomas, leiomyosarcomas, and benign metastasizing leiomyoma [8, 40–43]. Indeed, all the samples with 1p loss also displayed 19q loss. Interestingly, a recent study reported 1p/19q co-deletions in a subset of leiomyosarcomas lacking mutations in TP53 and RB1 [44]. In addition, 1p/19q co-deletions are common in oligodendrogliomas that harbor a mutation in isocitrate dehydrogenase 1 (IDH1) [45]. IDH1 and FH both encode for key enzymes of the citric acid cycle [46, 47]. We have previously shown UBE4B to be the most significantly downregulated gene on the 1p arm in leiomyomas harboring a 1p deletion [7]. UBE4B encodes for an additional conjugation factor, E4, which is involved in multiubiquitin chain assembly [48, 49]. It is tempting to speculate that the 1p/19q co-deletion may simultaneously target UBE2M on 19q13.43 and UBE4B on 1p36.22.
In this study, we have identified a novel uterine leiomyoma subtype with biallelic mutations in genes involved in neddylation of the Cullin 3-RING E3 ligase. Most mutated tumors displayed specific morphological features and a distinct gene expression profile characterized by activation of the NRF2 pathway. Compatible with functional studies on neddylation, our observations indicate disrupted neddylation as a novel mechanism leading to oncogenic NRF2 activation in human tumors. Development of NRF2 inhibitors has recently emerged as a promising anticancer strategy [50]. Overall, molecular stratification of uterine leiomyomas paves the way for more personalized treatment, as these very common tumors are still often considered as a single disease entity.
Materials and methods
Study material
The study has been approved by The Ethics Review Board of Hospital District of Helsinki and Uusimaa, Helsinki, Finland (HUS; 88/13/03/03/2015). Samples were collected with written informed consent from the patients or with authorization from the National Supervisory Authority of Welfare and Health (Valvira). The study material consisted of archival formalin-fixed paraffin-embedded (FFPE) tissue samples and corresponding hematoxylin-eosin (HE) stained slides that were obtained from the Department of Pathology, Helsinki University Hospital, Helsinki, Finland. Histopathological examination was performed by a pathologist specialized in gynecological pathology (RB or AP). The initial discovery set consisted of 141 leiomyomas with variant histopathology [18]. The validation set included 360 unselected leiomyomas, of which 12 displayed variant histopathology [20]. Three samples in the validation set were also included in the discovery set and therefore omitted from the latter cohort. The total sample series thus included 498 tumor samples, of which 348 displayed conventional and 150 variant histopathology. The status of MED12, HMGA2, and FH has been determined in previous studies [7, 18–20]. See Supplementary Table 1 for an overview of the samples. Normal tissue samples (myometrium or fallopian tube) were available for eight of the twelve patients with an AKR1B10hi tumor. In addition, an in-house exome-sequencing dataset of seven fresh frozen leiomyomas and five corresponding normal tissue samples was examined. FFPE tissue and corresponding HE-stained slides were obtained from one patient (1055) whose tumor showed two NAE1 mutations.
Immunohistochemistry
AKR1B10 immunohistochemistry was performed using tissue microarrays (TMA). When the sample showed positive staining in the TMA analysis or if the result was unclear, the corresponding whole tissue section was analyzed. The FH status for the variant samples has been previously determined with a 2SC antibody [7] or with an FH antibody [19]. In this study, samples previously stained only with the FH antibody were stained with the 2SC antibody for consistency. In brief, after deparaffination, heat-introduced antigen retrieval, and peroxidase blocking, incubation with the primary antibody AKR1B10 (1:300, H00057016-M01, Abnova, Taipei, Taiwan), 2SC (1:1000, crb2005017d, Discovery Antibodies, Billingham, UK), or NQO1 (1:500, sc-32793, Santa Cruz Biotechnology, Inc., Dallas, TX, USA) was performed overnight. Post antibody blocking (Immunologic BV, Duiven, Netherlands: post antibody blocking for bright vision plus) was followed by incubation with a secondary poly-HRP antibody (ImmunoLogic: Poly-HRP-GAM/R/R IgG). DAB Quanto (Thermo Fisher Scientific, Waltham, MA, USA) system was used to detect the expression levels. A pathologist (RB) evaluated the expression levels using a three-grade scoring system: ++ = strong staining, + = weak staining, and – = no staining. Both strong and weak staining were considered positive.
DNA and RNA extraction
Representative areas of tumor or normal tissue were marked on HE slides by a pathologist. Depending on the marking, DNA was extracted from the whole FFPE tissue sections or from six representative 0.8 mm cores with the ReliaPrep FFPE gDNA Miniprep System (Promega, Madison, WI, USA). DNA from FFPE samples that were selected for whole-exome (WES) and whole-genome sequencing (WGS) were extracted using the standard phenol-chloroform method. Fresh frozen tissue samples had been extracted with the FastDNA Spin Kit (MP Biomedicals, Santa Ana, CA, USA).
Total RNA was extracted and purified from macrodissected sections using the RNeasy® FFPE Kit (QIAGEN, Hilden, Germany) and the deparaffinization solution (QIAGEN) according to the manufacturer’s instructions. The concentration and purity of the extracted RNA were examined using the LabChip GX Touch HT RNA Assay Reagent Kit (PerkinElmer, Waltham, MA, USA) and the Qubit RNA BR kit (Thermo Fisher Scientific, Waltham, MA, USA). Genomic DNA contamination was measured with the Qubit DNA BR kit (Thermo Fisher Scientific).
Whole-exome and whole-genome sequencing
Tissue samples from 13 patients were subjected to WES and/or WGS. WES was performed with twelve tumors and two corresponding normal tissue samples at Biomedicum Functional Genomics Unit (FuGU), Helsinki, Finland on the Illumina (Illumina, San Diego, CA, USA) NextSeq 500 platform using 2 × 75bp paired-end reads. These 14 samples were prepared using the KAPA Hyper Prep kit (Roche NimbleGen, Madison, WI, USA) and captured using the SeqCap EZ MedExome Kit (Roche). WGS was performed with nine tumors and six corresponding normal tissue samples at Beijing Genomics Institute (BGI) on the BGISEQ-500 platform using 2 × 150 bp or 2 × 100 paired-end reads. These samples were prepared using the KAPA Hyper Prep kit (Roche). See Supplementary Table 7 for an overview of the methods that were used for each sample. One WES tumor was not analyzed for point mutations and microindels as it displayed very low quality and coverage.
Adapter and read trimming were performed using Trimmomatic [51]. WGS samples that were sequenced using 150bp reads were trimmed to 100 bp in length. WES and WGS data were preprocessed according to Genome Analysis ToolKit 4 best practices [52]. In brief, the samples were aligned against the Genome Reference Consortium Human Build 38 genome using BWA-MEM [53], duplicate reads were removed using Mark Duplicates, and base quality score recalibration was performed using BaseRecalibrator.
Paired and non-paired somatic variant calling was performed using Mutect2 with default parameters [52]. FFPE artifacts were identified using LearnReadOrientationModel [52]. Identified variants were filtered against a panel of normals consisting of 48 exomes and 28 genomes, a panel of normals generated from the 1000 genomes project, and variants present in the Genome Aggregation Database (exomes and genomes v2.0.1 and v3) using Baseplayer [54]. Recurrent nonsynonymous variants and microindels with a sequencing depth of at least 16, an allelic count of at least 8, and an allelic fraction of 0.3 were evaluated further using BasePlayer and annotated using variant effect predictor [55].
Somatic copy number alterations (SCNA) were called using CNVkit with default parameters [56]. SCNA for WGS samples were called against a pooled normal generated using six corresponding normal tissue samples, and SCNA for WES samples were called against a pooled normal generated using five in-house FFPE normal tissue samples. SCNA for one fresh frozen sample were called against a pooled normal generated using eight in-house fresh frozen normal tissue samples. Heatmaps of SCNA were visualized with the -d option to de-emphasize low-amplitude segments. WGS calls were used for visualization, unless only WES data was available.
Sanger sequencing
Mutations in UBE2M, NEDD8, CUL3, and NAE1 were validated by direct Sanger sequencing. One tumor that failed in exome sequencing was screened for mutations in the coding regions of UBE2M and NEDD8. Primers and conditions are available upon request. Sequencing was performed using the Applied Biosystems (ABI) 3730 DNA Sequencer at the Institute for Molecular Medicine Finland (FIMM), Helsinki, Finland. Sequence graphs were analyzed and visually inspected using Mutation Surveyor (SoftGenetics, State College, PA, USA) and FinchTV (Geospiza, Inc, Seattle, WA, USA).
3′RNA sequencing
3′RNA sequencing was performed with 13 leiomyomas and 7 corresponding myometrium samples as previously described [16]. These 20 samples were analyzed together with a previously published dataset of 44 leiomyomas (13 leiomyomas with a MED12 mutation, 15 with significant HMGA2 overexpression, and 16 with FH- deficiency) and 5 myometrium samples [16].
Dual-indexed mRNA libraries were prepared using QuantSeq 3′mRNA-Seq Library Prep Kit FWD (Lexogen Gmbh, Vienna, Austria) following the manufacturer’s protocol. Sequencing was performed at FIMM using the NovaSeq 6000 System (Illumina) with a read length of 2 × 101 base pairs and a minimum target coverage of 15 M reads for each library. FASTQ preprocessing was performed using the Integrated Data Analysis Pipeline version 2.3.1 FWD UMI (Lexogen Gmbh) implemented on the Bluebee® Genomics analysis platform. Briefly, the reads were trimmed with BBDuk, aligned to the Genome Reference Consortium human build 38 (GRCh38) reference genome using STAR [57], and counted using HTSeq [58]. Read counts of the technical replicates were merged. Raw read counts were normalized using DESeq2 [59]. Principal component analysis (PCA) and pairwise differential expression analysis were performed with DESeq2 on the Chipster platform [59, 60]. Pathway enrichment analysis of over-represented WikiPathways was performed using the WEB-based GEne SeT AnaLysis Toolkit [61].
TCGA and CCLE data analysis
TCGA mRNA counts were downloaded from the Broad GDAC Firehose database (2016_01_28) [62]. TCGA mutational data was obtained from the NCI Genomic Data Commons (v0.2.8.) [63]. Read counts were TMM-normalized and voom-transformed using edgeR (v3.26.8). To evaluate the performance of AKR1B10 expression as a classifier for NRF2 activation, we performed ROC analysis using ROCR (v1.0–11) [64]. Samples with truncating and well-established hotspot mutations in NFE2L2 and KEAP1 [65] served as positive controls for NRF2 activation. Identified mutations in UBE2M, NAE1, NEDD8, and CUL3 were visualized using maftools (R-version 3.6.0) [66]. Pre-normalized CCLE mRNA and proteomic data was downloaded from DepMap [67, 68] and CCLE (20Q4) [69] databases.
Gene expression microarray data analysis
Affymetrix GeneChip Human Genome U133 Plus 2.0 microarray data of melanoma cells that had been treated with MLN4924 or DMSO were obtained from the ArrayExpress database (accession: E-GEOD-30531). The data was normalized by the RMA method with affy (v1.62.0) R-package. Differential expression analysis was conducted using limma and EdgeR [70, 71]. Time course differential expression analysis was performed using limma. GSEA was performed with WikiPathways (Java GSEA v3.0) and the Singh NFE2L2 targets gene set, using MLN4924 treatment time as a continuous variable against DMSO treated samples [72, 73].
Supplementary information
Acknowledgements
We express gratitude to the patients for their contribution. We thank Lotta Katainen, Anne Mäkelä, and Maija Vahteristo for excellent technical assistance. We acknowledge CSC—IT Center for Science, Finland, for computational resources. Sequencing was performed at Biomedicum Functional Genomics Unit (FuGU) and at the Sequencing Unit of Institute for Molecular Medicine Finland (FIMM) Technology Centre, University of Helsinki, and at the Beijing Genomics Institute (BGI). This study was funded by Sigrid Jusélius Foundation, Academy of Finland (grants 307773 and 332697), Cancer Foundation Finland, the iCAN Digital Precision Cancer Medicine Flagship, Biomedicum Helsinki Foundation, UEF Doctoral Program of Molecular Medicine (DPMM), and Orion Research Foundation. Open access funded by Helsinki University Library.
Author contributions
P.V., M.M. and T.A. designed the study. R.B., A.P., A.Ä., P.P. and P.H. provided the samples. M.M., T.A., S.K. and S.R. performed the WES and WGS data analyses. M.M. performed the 3′RNA sequencing analysis. J.H. investigated TCGA and CCLE data. M.M. and J.H. analyzed the microarray data. T.A. carried out IHC staining and Sanger sequencing. R.B. re-evaluated the histopathology and scored IHC staining. T.H. and A-L.L. provided conceptual advice. M.M., T.A., P.V., S.K. and J.H. wrote the manuscript. P.V. supervised the study. All authors reviewed and approved the final manuscript.
Data availability
All key findings are presented in the manuscript and supplementary files. The raw data are not publicly available due to compliance with the ethics approval and confidentiality agreements. Data may be obtained from the authors upon reasonable request when compatible with European General Data Protection Regulation (GDPR) and with the permission from the University of Helsinki.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Miika Mehine, Terhi Ahvenainen.
Supplementary information
The online version contains supplementary material available at 10.1038/s41389-022-00425-3.
References
- 1.Stewart EA. Clinical practice. Uterine fibroids. N Engl J Med. 2015;372:1646–55. doi: 10.1056/NEJMcp1411029. [DOI] [PubMed] [Google Scholar]
- 2.Cardozo ER, Clark AD, Banks NK, Henne MB, Stegmann BJ, Segars JH. The estimated annual cost of uterine leiomyomata in the United States. Am J Obstet Gynecol. 2012;206:211.e211–9. doi: 10.1016/j.ajog.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ip PPC, Bennett JA, Croce S, Garg K, Yang B. Uterine leiomyoma. In: Rabban JT, Nucci MR, Malpica A, Brenton JD, Hui P, Ordi J et al., editors. WHO classification of tumors: female genital tumours. 5th edn. Lyon, France: IARC Publications; 2020. pp 272–6.
- 4.Mehine M, Mäkinen N, Heinonen HR, Aaltonen LA, Vahteristo P. Genomics of uterine leiomyomas: insights from high-throughput sequencing. Fertil Steril. 2014;102:621–9. doi: 10.1016/j.fertnstert.2014.06.050. [DOI] [PubMed] [Google Scholar]
- 5.Tomlinson IP, Alam NA, Rowan AJ, Barclay E, Jaeger EE, Kelsell D, et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet. 2002;30:406–10. doi: 10.1038/ng849. [DOI] [PubMed] [Google Scholar]
- 6.Berta DG, Kuisma H, Välimäki N, Räisänen M, Jäntti M, Pasanen A, et al. Deficient H2A.Z deposition is associated with genesis of uterine leiomyoma. Nature. 2021;596:398–403. doi: 10.1038/s41586-021-03747-1. [DOI] [PubMed] [Google Scholar]
- 7.Mäkinen N, Kämpjärvi K, Frizzell N, Bützow R, Vahteristo P. Characterization of MED12, HMGA2, and FH alterations reveals molecular variability in uterine smooth muscle tumors. Mol Cancer. 2017;16:101. doi: 10.1186/s12943-017-0672-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hodge JC, Pearce KE, Clayton AC, Taran FA, Stewart EA. Uterine cellular leiomyomata with chromosome 1p deletions represent a distinct entity. Am J Obstet Gynecol. 2014;210:572 e571–77. doi: 10.1016/j.ajog.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mehine M, Kaasinen E, Heinonen HR, Makinen N, Kampjarvi K, Sarvilinna N, et al. Integrated data analysis reveals uterine leiomyoma subtypes with distinct driver pathways and biomarkers. Proc Natl Acad Sci USA. 2016;113:1315–20. doi: 10.1073/pnas.1518752113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Podkalicka P, Mucha O, Kruczek S, Biela A, Andrysiak K, Stępniewski J, et al. Synthetically lethal interactions of heme oxygenase-1 and fumarate hydratase genes. Biomolecules. 2020;10:143. doi: 10.3390/biom10010143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song MY, Lee DY, Chun KS, Kim EH. The role of NRF2/KEAP1 signaling pathway in cancer metabolism. Int J Mol Sci. 2021;22:4376. doi: 10.3390/ijms22094376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ooi A, Wong JC, Petillo D, Roossien D, Perrier-Trudova V, Whitten D, et al. An antioxidant response phenotype shared between hereditary and sporadic type 2 papillary renal cell carcinoma. Cancer Cell. 2011;20:511–23. doi: 10.1016/j.ccr.2011.08.024. [DOI] [PubMed] [Google Scholar]
- 13.Pohl C, Dikic I. Cellular quality control by the ubiquitin-proteasome system and autophagy. Science. 2019;366:818–22. doi: 10.1126/science.aax3769. [DOI] [PubMed] [Google Scholar]
- 14.Rojo de la Vega M, Chapman E, Zhang DD. NRF2 and the Hallmarks of Cancer. Cancer Cell. 2018;34:21–43. doi: 10.1016/j.ccell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kerins MJ, Ooi A. A catalogue of somatic NRF2 gain-of-function mutations in cancer. Sci Rep. 2018;8:12846. doi: 10.1038/s41598-018-31281-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mehine M, Khamaiseh S, Ahvenainen T, Heikkinen T, Ayravainen A, Pakarinen P, et al. 3′RNA sequencing accurately classifies formalin-fixed paraffin-embedded uterine leiomyomas. Cancers (Basel). 2020;12:3839. doi: 10.3390/cancers12123839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahvenainen T, Kaukomaa J, Kämpjärvi K, Uimari O, Ahtikoski A, Mäkinen N, et al. Comparison of 2SC, AKR1B10, and FH antibodies as potential biomarkers for FH-deficient uterine leiomyomas. Am J Surg Pathol. 2022;46:537–46. doi: 10.1097/PAS.0000000000001826. [DOI] [PubMed] [Google Scholar]
- 18.Mäkinen N, Vahteristo P, Kämpjärvi K, Arola J, Bützow R, Aaltonen LA. MED12 exon 2 mutations in histopathological uterine leiomyoma variants. Eur J Hum Genet. 2013;21:1300–03. doi: 10.1038/ejhg.2013.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahvenainen TV, Mäkinen NM, von Nandelstadh P, Vahteristo MEA, Pasanen AM, Bützow RC, et al. Loss of ATRX/DAXX expression and alternative lengthening of telomeres in uterine leiomyomas. Cancer. 2018;124:4650–56. doi: 10.1002/cncr.31754. [DOI] [PubMed] [Google Scholar]
- 20.Äyräväinen A, Pasanen A, Ahvenainen T, Heikkinen T, Pakarinen P, Härkki P, et al. Systematic molecular and clinical analysis of uterine leiomyomas from fertile-aged women undergoing myomectomy. Hum Reprod. 2020;35:2237–44. doi: 10.1093/humrep/deaa187. [DOI] [PubMed] [Google Scholar]
- 21.Kopanos C, Tsiolkas V, Kouris A, Chapple CE, Albarca Aguilera M, Meyer R, et al. VarSome: the human genomic variant search engine. Bioinformatics. 2019;35:1978–80. doi: 10.1093/bioinformatics/bty897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47:D886–94. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng J, Guo J, Wang Z, North BJ, Tao K, Dai X, et al. Functional analysis of Cullin 3 E3 ligases in tumorigenesis. Biochim Biophys Acta Rev Cancer. 2018;1869:11–28. doi: 10.1016/j.bbcan.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou W, Xu J, Tan M, Li H, Li H, Wei W, et al. UBE2M is a stress-inducible dual E2 for neddylation and ubiquitylation that promotes targeted degradation of UBE2F. Mol Cell. 2018;70:1008–1024. e1006. doi: 10.1016/j.molcel.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Enchev RI, Schulman BA, Peter M. Protein neddylation: beyond cullin-RING ligases. Nat Rev Mol Cell Biol. 2015;16:30–44. doi: 10.1038/nrm3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baek K, Scott DC, Schulman BA. NEDD8 and ubiquitin ligation by cullin-RING E3 ligases. Curr Opin Struct Biol. 2020;67:101–9. doi: 10.1016/j.sbi.2020.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lo SC, Hannink M. CAND1-mediated substrate adaptor recycling is required for efficient repression of Nrf2 by Keap1. Mol Cell Biol. 2006;26:1235–44. doi: 10.1128/MCB.26.4.1235-1244.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wimuttisuk W, Singer JD. The Cullin3 ubiquitin ligase functions as a Nedd8-bound heterodimer. Mol Biol Cell. 2007;18:899–909. doi: 10.1091/mbc.e06-06-0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–6. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 30.Andérica-Romero AC, Hernández-Damián J, Vázquez-Cervantes GI, Torres I, González-Herrera IG, Pedraza-Chaverri J. The MLN4924 inhibitor exerts a neuroprotective effect against oxidative stress injury via Nrf2 protein accumulation. Redox Biol. 2016;8:341–7. doi: 10.1016/j.redox.2016.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou H, Lu J, Liu L, Bernard D, Yang CY, Fernandez-Salas E, et al. A potent small-molecule inhibitor of the DCN1-UBC12 interaction that selectively blocks cullin 3 neddylation. Nat Commun. 2017;8:1150. doi: 10.1038/s41467-017-01243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou H, Lu J, Chinnaswamy K, Stuckey JA, Liu L, McEachern D, et al. Selective inhibition of cullin 3 neddylation through covalent targeting DCN1 protects mice from acetaminophen-induced liver toxicity. Nat Commun. 2021;12:2621. doi: 10.1038/s41467-021-22924-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caballero OL, Resto V, Patturajan M, Meerzaman D, Guo MZ, Engles J, et al. Interaction and colocalization of PGP9.5 with JAB1 and p27(Kip1) Oncogene. 2002;21:3003–10. doi: 10.1038/sj.onc.1205390. [DOI] [PubMed] [Google Scholar]
- 34.Dubiel W, Chaithongyot S, Dubiel D, Naumann M. The COP9 Signalosome: a multi-DUB complex. Biomolecules. 2020;10:1082. doi: 10.3390/biom10071082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wada H, Kito K, Caskey LS, Yeh ET, Kamitani T. Cleavage of the C-terminus of NEDD8 by UCH-L3. Biochem Biophys Res Commun. 1998;251:688–92. doi: 10.1006/bbrc.1998.9532. [DOI] [PubMed] [Google Scholar]
- 36.Bishop P, Rocca D, Henley JM. Ubiquitin C-terminal hydrolase L1 (UCH-L1): structure, distribution and roles in brain function and dysfunction. Biochem J. 2016;473:2453–62. doi: 10.1042/BCJ20160082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miettinen M, Felisiak-Golabek A, Wasag B, Chmara M, Wang Z, Butzow R, et al. Fumarase-deficient uterine leiomyomas: an immunohistochemical, molecular genetic, and clinicopathologic study of 86 cases. Am J Surg Pathol. 2016;40:1661–9. doi: 10.1097/PAS.0000000000000703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanz-Ortega J, Vocke C, Stratton P, Linehan WM, Merino MJ. Morphologic and molecular characteristics of uterine leiomyomas in hereditary leiomyomatosis and renal cancer (HLRCC) syndrome. Am J Surg Pathol. 2013;37:74–80. doi: 10.1097/PAS.0b013e31825ec16f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Christacos NC, Quade BJ, Dal Cin P, Morton CC. Uterine leiomyomata with deletions of Ip represent a distinct cytogenetic subgroup associated with unusual histologic features. Genes Chromosomes Cancer. 2006;45:304–12. doi: 10.1002/gcc.20291. [DOI] [PubMed] [Google Scholar]
- 40.Quade BJ, Wang TY, Sornberger K, Dal Cin P, Mutter GL, Morton CC. Molecular pathogenesis of uterine smooth muscle tumors from transcriptional profiling. Genes Chromosomes Cancer. 2004;40:97–108. doi: 10.1002/gcc.20018. [DOI] [PubMed] [Google Scholar]
- 41.Levy B, Mukherjee T, Hirschhorn K. Molecular cytogenetic analysis of uterine leiomyoma and leiomyosarcoma by comparative genomic hybridization. Cancer Genet Cytogenet. 2000;121:1–8. doi: 10.1016/S0165-4608(00)00225-9. [DOI] [PubMed] [Google Scholar]
- 42.Bowen JM, Cates JM, Kash S, Itani D, Gonzalez A, Huang D, et al. Genomic imbalances in benign metastasizing leiomyoma: characterization by conventional karyotypic, fluorescence in situ hybridization, and whole genome SNP array analysis. Cancer Genet. 2012;205:249–54. doi: 10.1016/j.cancergen.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nucci MR, Drapkin R, Dal Cin P, Fletcher CD, Fletcher JA. Distinctive cytogenetic profile in benign metastasizing leiomyoma: pathogenetic implications. Am J Surg Pathol. 2007;31:737–43. doi: 10.1097/01.pas.0000213414.15633.4e. [DOI] [PubMed] [Google Scholar]
- 44.Williams EA, Sharaf R, Decker B, Werth AJ, Toma H, Montesion M, et al. CDKN2C-null leiomyosarcoma: a novel, genomically distinct class of TP53/RB1-wild-type tumor with frequent CIC genomic alterations and 1p/19q-codeletion. JCO Precis Oncol. 2020;4:PO.20.00040. doi: 10.1200/PO.20.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, et al. WHO classification of tumours of the central nervous system. 4th edn. Lyon: International Agency For Research On Cancer; 2016.
- 46.Grassian AR, Parker SJ, Davidson SM, Divakaruni AS, Green CR, Zhang X, et al. IDH1 mutations alter citric acid cycle metabolism and increase dependence on oxidative mitochondrial metabolism. Cancer Res. 2014;74:3317–31. doi: 10.1158/0008-5472.CAN-14-0772-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Woods SA, Schwartzbach SD, Guest JR. Two biochemically distinct classes of fumarase in Escherichia coli. Biochim Biophys Acta. 1988;954:14–26. doi: 10.1016/0167-4838(88)90050-7. [DOI] [PubMed] [Google Scholar]
- 48.Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96:635–44. doi: 10.1016/S0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- 49.Hoppe T, Matuschewski K, Rape M, Schlenker S, Ulrich HD, Jentsch S. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell. 2000;102:577–86. doi: 10.1016/S0092-8674(00)00080-5. [DOI] [PubMed] [Google Scholar]
- 50.Telkoparan-Akillilar P, Panieri E, Cevik D, Suzen S, Saso L. Therapeutic targeting of the NRF2 signaling pathway in cancer. Molecules. 2021;26:1417.. doi: 10.3390/molecules26051417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Katainen R, Donner I, Cajuso T, Kaasinen E, Palin K, Mäkinen V, et al. Discovery of potential causative mutations in human coding and noncoding genome with the interactive software BasePlayer. Nat Protoc. 2018;13:2580–600. doi: 10.1038/s41596-018-0052-3. [DOI] [PubMed] [Google Scholar]
- 55.McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, et al. The ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol. 2016;12:e1004873. doi: 10.1371/journal.pcbi.1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kallio MA, Tuimala JT, Hupponen T, Klemelä P, Gentile M, Scheinin I, et al. Chipster: user-friendly analysis software for microarray and other high-throughput data. BMC Genomics. 2011;12:507. doi: 10.1186/1471-2164-12-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013;41:W77–83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Broad Institute TCGA Genome Data Analysis Center: Firehose stddata__2016_01_28 run. Broad Institute of MIT and Harvard. 10.7908/C11G0KM9. http://gdac.broadinstitute.org/runs/stddata__2016_01_28/data/. 2016.
- 63.Grossman RL, Heath AP, Ferretti V, Varmus HE, Lowy DR, Kibbe WA, et al. Toward a shared vision for cancer genomic data. N. Engl J Med. 2016;375:1109–12. doi: 10.1056/NEJMp1607591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21:3940–41. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- 65.Chang MT, Bhattarai TS, Schram AM, Bielski CM, Donoghue MTA, Jonsson P, et al. Accelerating discovery of functional mutant alleles in cancer. Cancer Discov. 2018;8:174–83. doi: 10.1158/2159-8290.CD-17-0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28:1747–56. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet. 2017;49:1779–84. doi: 10.1038/ng.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, et al. Defining a cancer dependency map. Cell. 2017;170:564–576.e516. doi: 10.1016/j.cell.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nusinow DP, Szpyt J, Ghandi M, Rose CM, McDonald ER, 3rd, Kalocsay M, et al. Quantitative proteomics of the cancer cell line encyclopedia. Cell. 2020;180:387–402.e316. doi: 10.1016/j.cell.2019.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martens M, Ammar A, Riutta A, Waagmeester A, Slenter DN, Hanspers K, et al. WikiPathways: connecting communities. Nucleic Acids Res. 2021;49:D613–21. doi: 10.1093/nar/gkaa1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Singh A, Boldin-Adamsky S, Thimmulappa RK, Rath SK, Ashush H, Coulter J, et al. RNAi-mediated silencing of nuclear factor erythroid-2-related factor 2 gene expression in non-small cell lung cancer inhibits tumor growth and increases efficacy of chemotherapy. Cancer Res. 2008;68:7975–84. doi: 10.1158/0008-5472.CAN-08-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All key findings are presented in the manuscript and supplementary files. The raw data are not publicly available due to compliance with the ethics approval and confidentiality agreements. Data may be obtained from the authors upon reasonable request when compatible with European General Data Protection Regulation (GDPR) and with the permission from the University of Helsinki.