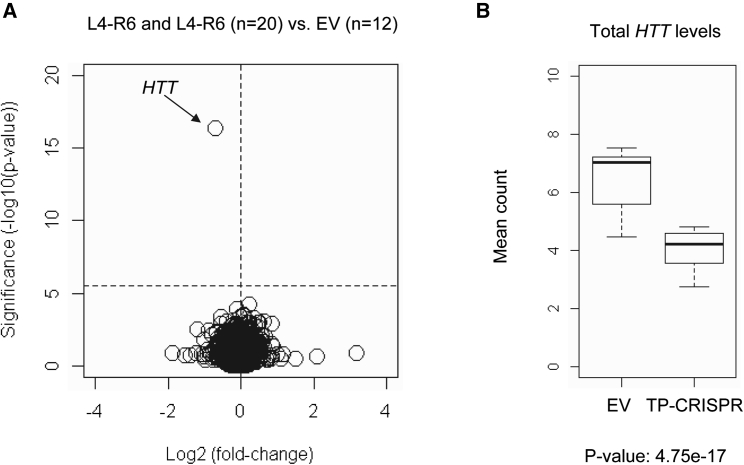
Figure 4.
On-target gene specificity supported by RNA-seq analysis
To increase the sensitivity and the power to detect any small but significantly altered genes, we combined 10 targeted clonal lines targeted by L4-R4 and 10 clonal lines targeted by L4-R6 gRNA combinations to be compared with 12 EV-treated controls. (A) Significance values (uncorrected p values on the y axis) were compared with log2 (fold-change) (x axis) to highlight significantly altered genes. A horizontal and a vertical line represent Bonferroni-corrected significance and zero fold-change, respectively. (B) Expression levels of total HTT (mutant plus normal) based on the DGE analysis are summarized in a boxplot. Total HTT levels were decreased by 38% in TP-CRISPR targeted clonal lines.