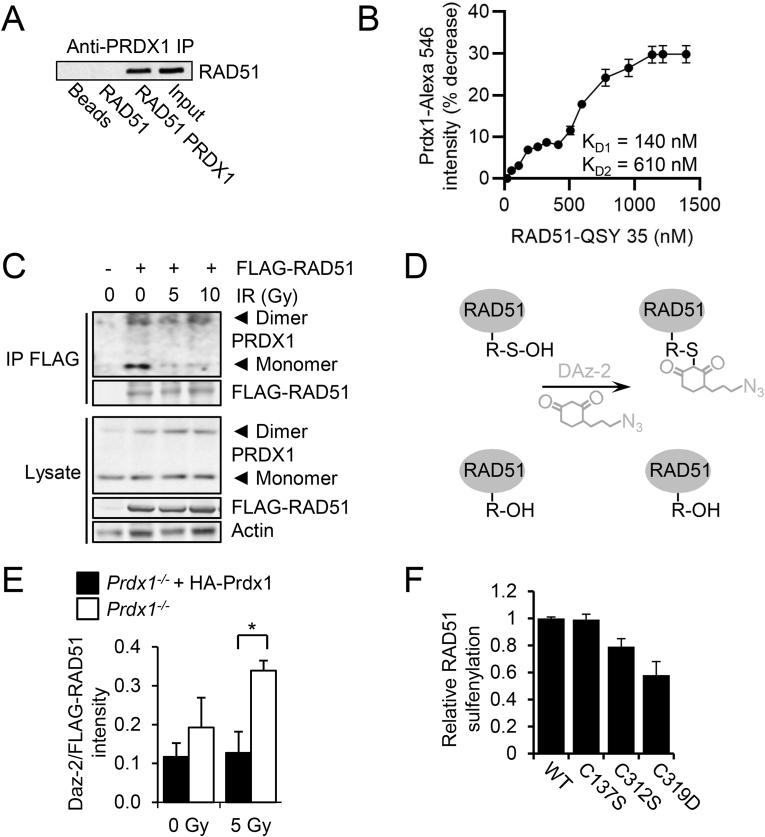
Fig. 3.
PRDX1 binds RAD51 and loss of PRDX1 increases RAD51 sulfenylation.
(A) In vitro co-IP reactions were performed with purified RAD51 and PRDX1 proteins by precipitating PRDX1 with antibody and immunoblotting for RAD51.
(B) Alexa Fluor 546 labelled PRDX1 was incubated with increasing concentrations of RAD51 labelled with the QSY35 quencher (0–1600 nM) to produce a binding curve where the decease of signal was used for KD quantification.
(C) FLAG immunoprecipitation of 293T cells demonstrated that endogenous dimeric and monomeric PRDX1 could bind FLAG-RAD51 when probed under non-reducing conditions. 293T cells were transfected with FLAG-RAD51 and irradiated with 0–10 Gy.
(D) DAz-2 reaction scheme.
(E) Prdx1−/− MEFs transduced with FLAG-RAD51 and HA-Prdx1 exhibited decreased sulfenylation as measured by DAz-2 incorporation. MEFs were treated with DAz-2 (5 mM) for 1.5 h and DAz-2 incorporation was detected by chemiluminescence by utilizing ligation of p-biotin and binding of streptavidin-HRP. Mean ± SEM as indicated.
(F) Prdx1−/− MEFs transduced with WT or cysteine mutant RAD51 expression constructs exhibited decreased sulfenylation by DAz-2 incorporation in RAD51C319 mutant samples. MEFs transduced with a RAD51 expression construct were treated with DAz-2 (5 mM) for 1.5 h. DAz-2 incorporation was detected by chemiluminescence by utilizing ligation of p-biotin and binding of streptavidin-HRP. The mean and range of two experiments is indicated.