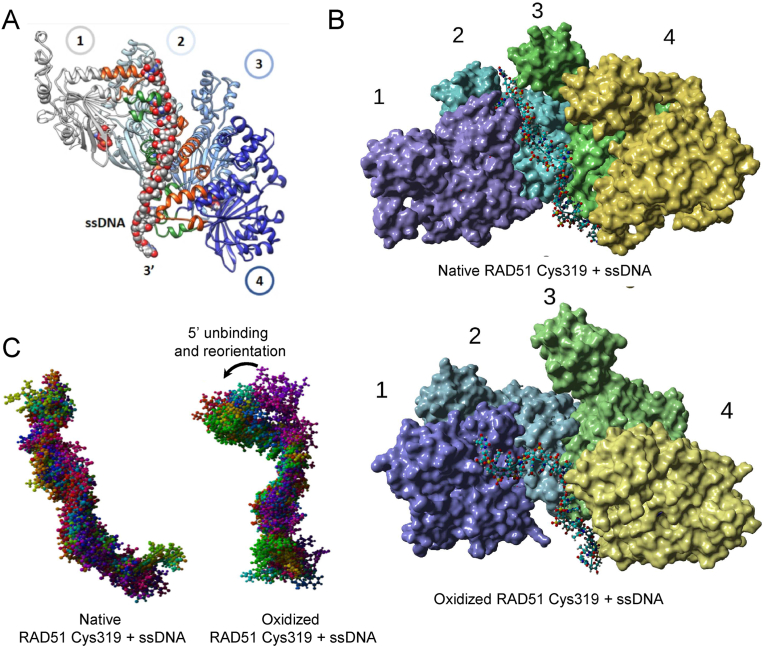
Fig. 4.
MD simulations of a tetrameric RAD51-DNA filament show sulfenylation of Cys319 destabilizes ssDNA binding.
(A) The model was designed by superposing our previously published all atom homology model [32] onto a tetrameric cryo-EM structure of RAD51 (5NP7), and subsequently superposing poly-dT from a RecA (3CMU) structure. This homology model was a starting point for an MD-based refinement, and subsequent MD simulations comparing native and Cys319 oxidized filaments. RAD51 protomers are labelled 1–4 with ssDNA.
(B) The last snap shot of a refined MD for native (upper) and oxidized (lower) RAD51 filaments with different protomers colored in purple, blue, green and yellow demonstrated ssDNA (poly-dT ball and stick) was destabilized in the oxidized RAD51 filament from binding residues in loops 232–237 and 278–288 present in the native RAD51 Cys319 filament.
(C) Superposed DNA snapshots without RAD51 filaments shown revealed that the native RAD51 filament yielded stable DNA positioning in an extended conformation throughout the simulation (left), while the RAD51 filament with oxidized Cys319 residues exhibited a complex behavior where the 5′ region of the ssDNA initially became unbound from monomer 1 and 2 (purple) and then migrates over the course of the simulation to bind the N-terminal domain of monomer 1 (green). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)