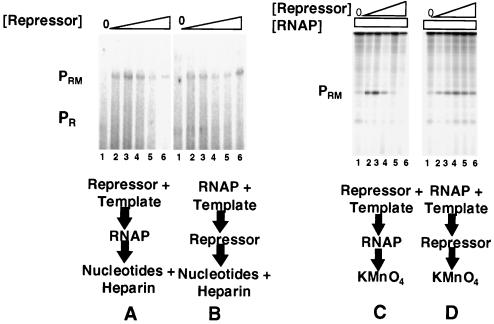
FIG. 7.
Damaging PR obviates RNA polymerase inhibition of repressor-activated PRM transcription (A and B) or open complex formation (C and D). For panels A and B, DNA fragments containing wild-type PRM and −10 mutant PR (see Fig. 1 for sequence) were transcribed in vitro in the absence of repressor (lanes 1) and at various increasing repressor concentrations. Repressor concentrations were increased in 2.5-fold steps starting at 250 nM protein (lanes 2 to 6). Positions of transcripts resulting from initiation of transcription from PRM and PR are indicated. (A) The 434 repressor was incubated with DNA template at 23°C for 10 min, followed by addition of RNA polymerase. The reaction mixture was transferred to 37°C for 10 min before the transcription reaction was initiated by the addition of nucleotides and heparin. (B) RNA polymerase was incubated with DNA at 37°C for 10 min before addition of the 434 repressor. After an additional 10-min incubation at 37°C, the transcription reaction was initiated by the addition of nucleotides and heparin. (C and D) Shown are the open complexes formed at PRM in the absence of the repressor (lanes 1) and the presence of increasing concentrations of the 434 repressor as detected by KMnO4 footprinting. Repressor concentrations were increased in 2.5-fold steps starting at 250 nM protein. Footprinting conditions are given in Materials and Methods. Positions of PR and PRM open complex are indicated. (C) Incubation conditions were as described for panel A. (D) Incubation conditions were as described for panel B. RNAP, RNA polymerase.