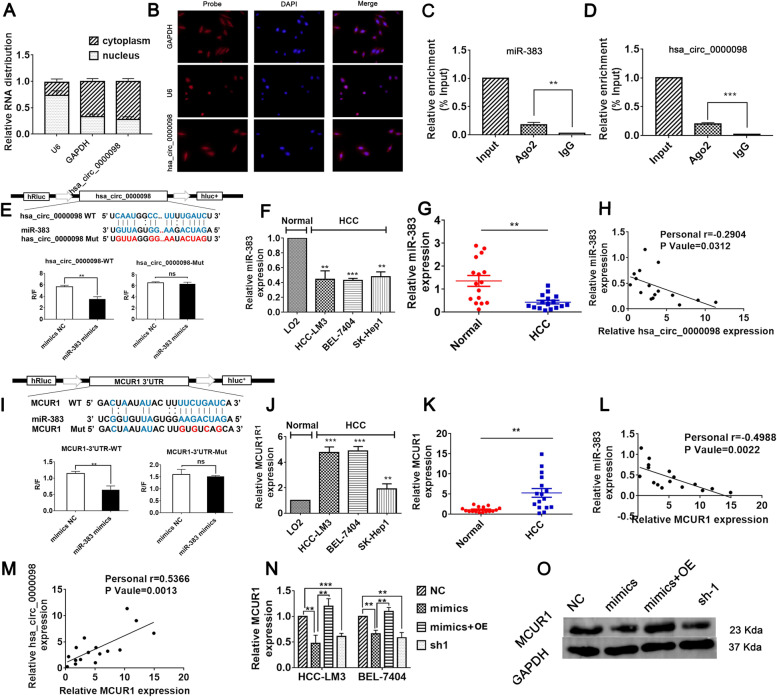
Fig. 3.
Circ_0000098 regulates MCUR1 expression by sponging miR-383. A The cytoplasmic and nuclear levels of circ_0000098 in HCC-LM3 cells were examined using RT-qPCR assays. GAPDH and U6 were utilized as cytoplasmic and nuclear controls, respectively. B FISH assay examined the localization of circ_0000098 in HCC-LM3 cells. GAPDH and U6 were utilized as cytoplasmic and nuclear controls, respectively. C, D The Ago2 RIP assay was used to evaluate miR-383 (C) and circ_0000098 (D) expression in HCC-LM3 cells. E Bioinformatics prediction of the binding sites between circ_0000098 and miR-383. Wild-type (WT) and mutant (Mut) binding sequences for circ_0000098 were cloned into a luciferase reporter. Relative luciferase activity (R/F) was analyzed for HCC-LM3 cells co-transfected with miR-383 mimics, control mimics (mimics NC), along with WT or Mut luciferase reporter vectors. F Detection of miR-383 expression in HCC cell lines and LO2 cells by RT-qPCR. G Examination of miR-383 expression within HCC and adjacent normal tissues by RT-qPCR. H Correlation analyses of miR-383 and circ_0000098 expression in HCC tissues. I Bioinformatics predictions for binding sites between miR-383 and MCUR1 3′-UTR. The WT and Mut binding sequences of MCUR1 3′-UTR were cloned into the luciferase reporter. R/F was analyzed in HCC-LM3 cells co-transfected with miR-383 mimics or mimics NC, together with WT or Mut luciferase reporter vectors. J Detection of MCUR1 mRNA expression in HCC cells and LO2 cells using RT-qPCR assays. K Detection of MCUR1 mRNA expression in HCC and adjacent normal tissues by RT-qPCR. L, M Correlation analyses of miR-383 and MCUR1 mRNA expression (L) and circ_0000098 and MCUR1 mRNA levels (M) in HCC tissues. N Detection of MCUR1 mRNA expression in HCC cells transfected as indicated using RT-qPCR assays. O Western blotting analysis of MCUR1 expression in the indicated cells. **P < 0.01, ***P < 0.001, NS: not significant