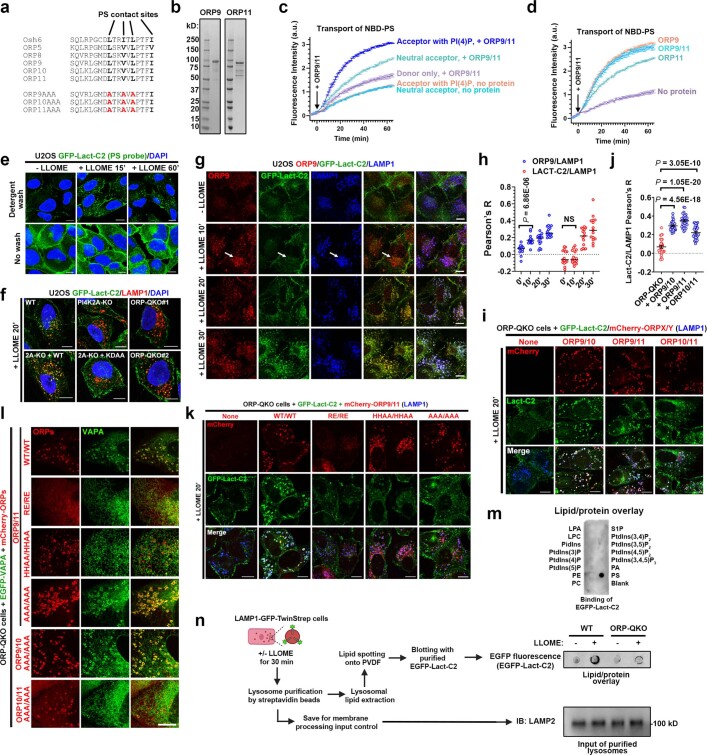
Extended Data Fig. 7. ORP9/10/11 transport phosphatidylserine (PS) to damaged lysosomes.
a, Alignment of human ORPs with yeast Osh6 which specifically transports phosphatidylserine (PS) but not sterols. Bold residues are direct PS contact sites of Osh6. The AAA mutants are designed to lose PS-binding, which have been confirmed in this study. b, Coomassie blue staining of purified ORP9 and ORP11. c, FRET-based assay demonstrating higher lipid transport activity by ORP9/11 (50 nM/50 nM = 100 nM in total) in the presence of PtdIns4P in the acceptor liposome. Note that there was a slight increase in NBD fluorescence in the absence of acceptor liposomes (donor only), which is consistent with ORP9/11-mediated NBD-PS extraction from the donor membranes without further delivery to acceptor membranes. d, ORP9/11 monomers and heterodimers show similar PS transport activity in vitro. ORP9/11 (50 nM/50 nM) heterodimers or monomers (100 nM each) were added to donor and acceptor liposome mixtures, and the changes in NBD fluorescence was monitored over time. The acceptor liposomes in all reactions contained 5% PtdIns4P. See also Supplementary Results. e, The PS probe GFP-Lact-C2 is quickly recruited to damaged lysosomes. U2OS cells stably expressing GFP-Lact-C2 were treated with 1 mM LLOME for the indicated time periods and then briefly washed with 0.1% Triton-X100 (detergent wash, see Methods) to remove background PS signals from the cytosol. The cells were then immediately fixed. f, The recruitment of GFP-Lact-C2 to damaged lysosomes is dependent on PI4K2A and its kinase activity, as well as the ORPs enriched on damaged lysosomes. Cells were treated with LLOME and detergent-washed before fixation and immunostaining. The colocalization of GFP-Lact-C2 and LAMP1 was quantified in Fig. 3d. g, ORP9 is recruited to damaged lysosomes earlier than the PS probe GFP-Lact-C2. U2OS Cells stably expressing GFP-Lact-C2 were treated with LLOME and detergent-washed before fixation and immunostaining of endogenous ORP9 and LAMP1. Arrow indicates ORP9 puncta negative for GFP-Lact-C2. h, The colocalization between LAMP1 and ORP9 or GFP-Lact-C2 in (g) was quantified. Data show mean ± sem of Pearson’s correlation coefficient; n = 15 cells over three trials for each condition. i, LMP-induced lysosomal PS transport in ORP-QKO cells can be rescued by the reconstitution of any two of ORP9/10/11. Cells were treated with 1 mM LLOME for 20 min and detergent-washed before being fixed for immunostaining of LAMP1. j, The colocalization of GFP-Lact-C2 and LAMP1 in (i) was quantified. Data show mean ± sem of Pearson’s correlation coefficient; n = 30 cells for each condition. k, ORP-QKO cells stably expressing GFP-Lact-C2 and mCherry-ORP9/11 or their mutants were treated with LLOME for 20 min and detergent-washed before being fixed for immunostaining of LAMP1. See Fig. 3k for quantification. l, The LMP-induced lysosomal recruitment of VAPA in ORP-QKO cells can be strongly rescued by the AAA mutants (loss of PS binding to the lipid transport domain) of ORP9/10 or ORP9/11, weakly rescued by ORP10/11 or the HHAA mutants of ORP9/11, and cannot be rescued by the RE mutants of ORP9/11. See Fig. 3e for details of the mutants. ORP-QKO cells stably expressing EGFP-VAPA and the indicated mCherry-ORP proteins were treated with LLOME for 10 min before fixed for confocal microscopy. m, PIP strips assay showing specific binding of purified EGFP-Lact-C2 to PS. n, Schematic illustration for lysosomal purification, lipid extraction, PVDF spotting, and PS detection. Wild type or ORP-QKO cells stably expressing LAMP1-GFP-twin-Strep were treated or not with 1 mM LLOME for 30 min, followed by lysosomal purification using sptreptavidin beads. Lipids were extracted from lysosomes on beads and dropped onto PVDF membrane for PS detection by purified EGFP-Lact-C2. On the right are representative images of the lipid/protein overlay assay for lysosomal PS detection with LAMP2 immunoblots as total membrane input controls. DAPI stains the nuclei. Bar, 10 μm. NS, not significant. Uncropped western blot images are provided in Supplementary Fig. 1. Source data for graphs in this Figure are provided.