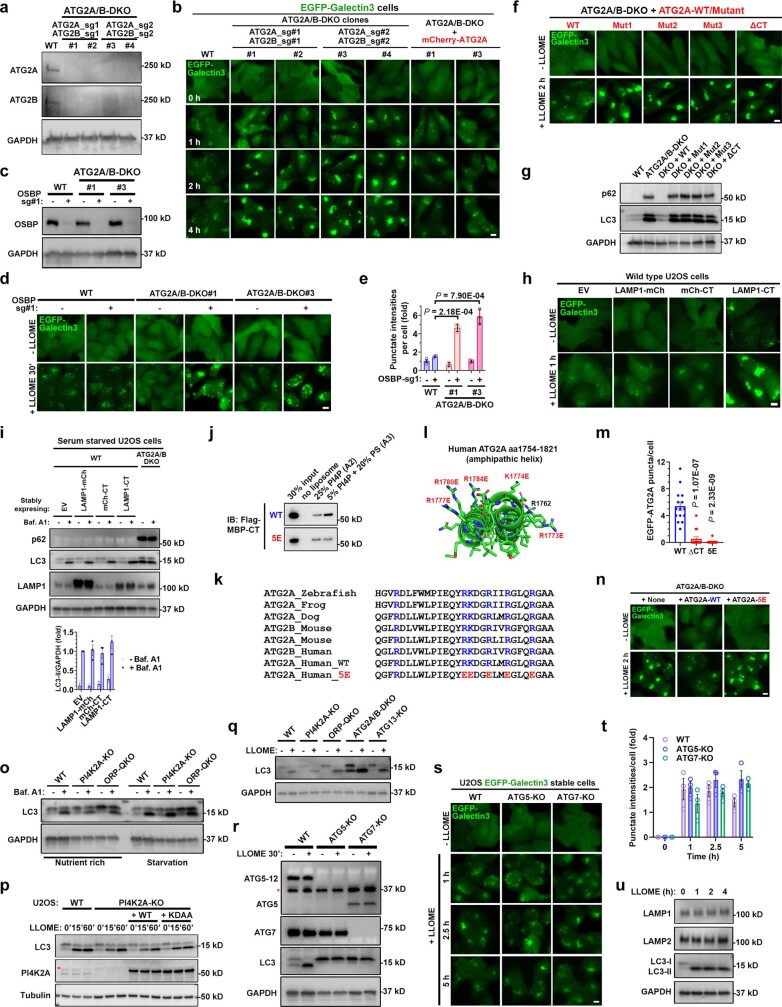
Extended Data Fig. 9. Independent of macroautophagy, ATG2 mediates rapid lysosomal repair through its lipid transport activity stimulated by PS.
a, Immunoblotting of ATG2A/B protein levels in wild type or ATG2A/B-DKO cells. Two individual clones from each set of CRISPR guides were used for further characterization. b, Double knockout of ATG2A/B causes robust defects of rapid lysosomal repair as shown by the EGFP-Galectin3 assay. The same knockout clones from (a) were used for this assay. Cells were continuously challenged with LLOME and live cell images were captured at indicated time points. Note that re-expression of ATG2A was sufficient to rescue rapid lysosomal repair in the DKO cells. See Fig. 4c for quantifications. c, Immunoblotting of OSBP levels in indicated CRISPR pools. d, Further deletion of OSBP in ATG2A/B-DKO cells causes dramatic defects in rapid lysosomal repair at an early time point. Note that loss of OSBP or ATG2A/B alone does not cause apparent defects within 30 min of LLOME treatment, indicating functional redundancy. e, quantification of the early time point Galectin3 intensities above threshold in (d). About 50–100 random cells were quantified for each condition. Mean ± sem; n = 3. f, Four distinct ATG2A lipid transport mutants are unable to rescue rapid lysosomal repair in ATG2A/B-DKO cells. Stable U2OS cell lines with indicated genetic modifications were continuously challenged with LLOME and live cell images were captured at indicated time points. See quantification in Fig. 4h. g, The ATG2A lipid transport mutants cannot restore autophagic turnover in ATG2A/B-DKO cells. Stable U2OS cell lines with indicated genetic modifications were directly harvested for whole cell lysate extraction followed by immunoblotting of indicated proteins. Data represent more than five experiments. h, EGFP-Galectin3 assay showing the defects of rapid lysosomal repair in wild type cells stably expressing LAMP1-CT but not mCherry-CT or LAMP1-mCherry. See quantification in Fig. 4j. i. Overexpression of different ATG2A-CT fusion proteins including LAMP1-CT does not block macroautophagy. Cells with indicated genetic modifications were treated with 100 nM Bafilomycin A1 for 4 h, followed by immunoblotting of indicated proteins. ATG2A/B-DKO cells served as a positive control for autophagy defects with marked accumulation of both p62 and LC3-II. Quantification of LC3-II intensities normalized to GAPDH is shown. Mean ± sem; n = 3. j, Liposome pull down assays testing the membrane binding capacity of purified MBP–CT or its 5E mutant. k, Highly conserved basic residues in ATG2A-CT. The residues mutated in 5E are in red and also shown in panel (l). l, AlphaFold structure of ATG2A-CT (amino acids 1754–1821). Six highly conserved basic residues are labeled, five of which were mutated in the 5E mutant. m, ATG2A-ΔCT and −5E mutants form dramatically reduced numbers of puncta in response to lysosomal damage. U2OS cells stably expressing EGFP-tagged ATG2A-WT, -ΔCT or −5E were stimulated with LLOME and the numbers of EGFP-ATG2A puncta 20 min after stimulation were determined using live cell imaging. Mean ± sem; n = 15 cells over three trials for each condition. n, EGFP-Galectin3 assay showing the failure of ATG2A-5E mutant in rescuing rapid lysosomal repair in ATG2A/B DKO cells. See quantification in Fig. 4m. o, Knockout of PI4K2A or ORPs does not affect LC3 turnover, indicating normal macroautophagy. Indicated cell lines were treated with 100 nM bafilomycin A1 for four hours followed by whole cell lysate harvest for immunoblotting. p, PI4K2A activity does not affect LLOME-induced LC3 lipidation. U2OS cells with indicated genetic modifications were treated with 1 mM LLOME for 15 to 60 min and then whole cell lysates were analyzed for the level of LC3. Asterisk indicates a nonspecific band. q, LLOME-induced LC3 lipidation is independent of PI4K2A, ORPs, ATG2, and ATG13. U2OS cells with indicated genetic modifications were treated with 1 mM LLOME for 30 min and then whole cell lysates were analyzed for the level of LC3. r, Immunoblotting shows the loss of ATG5 and ATG7 in relevant U2OS knockout cells. Asterisk indicates a nonspecific band. Note that ATG5 is conjugated to ATG12 in wild type cells, which shows a higher band compared with unconjugated ATG5 in ATG7-KO cells. s, ATG5-KO and ATG7-KO cells have normal rapid lysosomal repair, without increased EGFP-Galectin 3 puncta after continuous challenging with 1 mM LLOME. Bar, 10 μm. t, Quantification of EGFP-Galectin3 intensities above threshold in (s). 50–100 random cells were quantified for each condition. Mean ± sem; n = 3. u, Immunoblotting shows no evidence of degradation of either LAMP1 or LAMP2 within four hours of LLOME treatment. U2OS cells were treated with 1 mM LLOME for 1 to 4 h and whole cell lysates was then analyzed for levels of the indicated protein. DAPI stains the nuclei. Bar, 10 μm. Uncropped western blot images are provided in Supplementary Fig. 1. Source data for graphs in this Figure are provided.