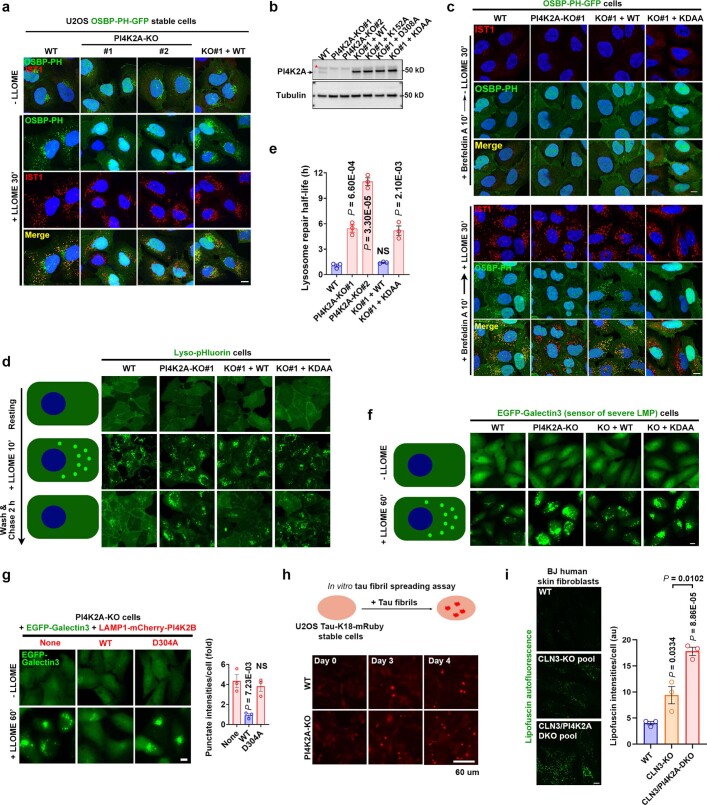
Extended Data Fig. 4. PI4K2A-meidated lysosomal PtdIns4P signaling is essential for rapid lysosomal repair.
a, PI4K2A is required for PtdIns4P production on damaged lysosomes. Wild type (WT) or two independent PI4K2A-KO cell lines stably expressing the PtdIns4P probe OSBP-PH-GFP were treated with or without LLOME for 30 min before being fixed for immunostaining of endogenous IST1, a marker for damaged lysosomes. The images show the loss of LMP-induced lysosomal recruitment of OSBP-PH-GFP in PI4K2A-KO cells, which is fully rescued by re-expression of WT PI4K2A. Note, some green puncta still remain in PI4K2A-KO cells, corresponding to the Golgi pool of PtdIns4P. These puncta are absent after Brefeldin A treatment. See panel (c) and Fig. 1g. Bar, 10 μm. b, Western blot verification of PI4K2A levels in wild type or genetically modified U2OS cells. Asterisk indicates a nonspecific band. c, PI4K2A recruitment is responsible for PtdIns4P production on damaged lysosomes. OSBP-PH-GFP-expressing U2OS cells with the indicated genetic modifications were pretreated for 10 min with Brefeldin A to disassemble the Golgi, removing background GFP puncta on the Golgi complex. Cells were then treated with or without 1 mM LLOME for 30 min before immunostaining of endogenous IST1. See quantification in Fig. 1g. Bar, 10 μm. d, The Lyso-pHluorin assay reveals the requirement of PI4K2A and its kinase activity in rapid lysosome repair. Schematic illustration of the assay is shown on the left. Lyso-pHluorin was stably expressed in various U2OS cells with indicated genetic modifications. Cells were treated with 1 mM LLOME for 10 min and then gently washed twice with pre-warmed culture media; fluorescent images were taken at the indicated time points. See quantification in Fig. 1h. Bar, 10 μm. e, Quantification of the lysosomal repair half-life of different cell lines using the Lyso-pHluosin assay. Data were derived from the experiment in (d). Mean ± sem; n = 3. f, The EGFP-Galectin3 rapid lysosomal repair assay demonstrates a role for PI4K2A in protecting lysosomes from severe damage. Schematic illustration of the assay is shown on the left. U2OS cells stably expressing EGFP-Galectin3 were treated with 1 mM LLOME for 60 min to continuously damage lysosomes. Fluorescence microscopic images were taken using live cells. Note, brighter Galectin3 puncta indicates larger and greater unrepaired lysosomal membrane pores. See quantification in Fig. 1i. Bar, 10 μm. g, Rapid lysosomal repair in PI4K2A-KO cells is rescued by expressing LAMP1-mCherry-PI4K2B but not its kinase dead mutant. Cells stably expressing indicated proteins were treated as in (f) and EGFP-Galectin3 punctate intensities were quantified. Mean ± sem; n = 3. Bar, 10 μm. h, Knockout of PI4K2A accelerates tau fibril spreading in cell culture. Top, Schematic illustration of the tau fibril spreading assay. Bottom, cells were incubated with sonicated, non-fluorescent tau fibrils. The percentage of cells showing bright tau-K18-mRuby puncta were examined on a daily basis. See quantification in Fig. 1j. Bar, 60 μm. i, Deletion of PI4K2A exacerbates lipofuscin accumulation in CLN3-knockout fibroblasts. Left, BJ human skin fibroblasts were infected with CRISPR lentivirus for the deletion of CLN3 alone or double deletion of PI4K2A and CLN3. CLN3-sg#1 and PI4K2A-sg#2 were used to obtain the knockout pools. Six days after infection, cells were fixed directly for lipofuscin detection. Right, quantification of lipofuscin intensities. 10–20 random cells per condition; Mean ± sem; n = 3. Bar, 10 μm. DAPI stains the nuclei. NS, not significant. Uncropped western blot images are provided in Supplementary Fig. 1. Source data for graphs in this Figure are provided.