Figure 3.
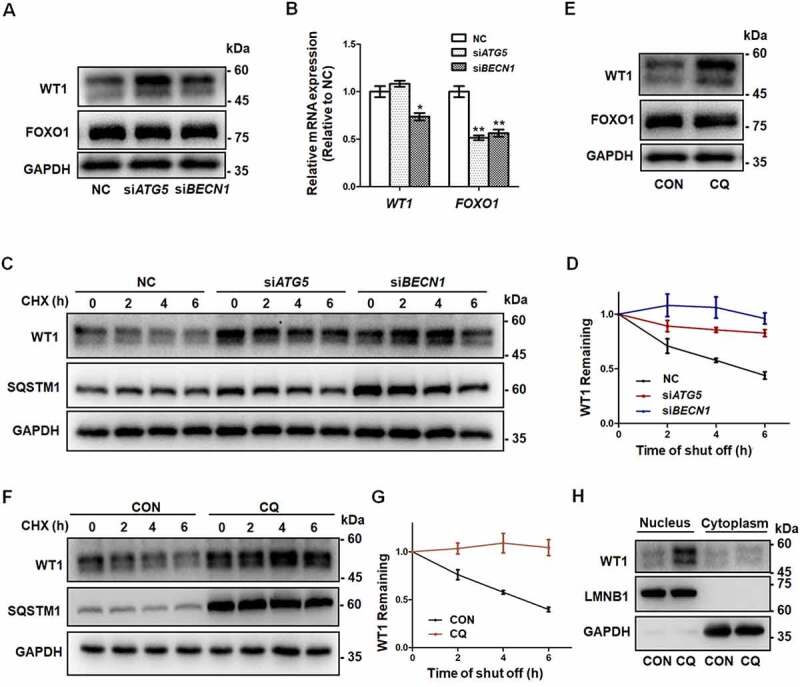
Accumulation of WT1 in GCs results from autophagy inhibition. (A) Western blot showed that, compared to NC, there was an increase in WT1 protein in both the siATG5 and siBECN1 groups, but there were no significant changes in FOXO1 protein. GAPDH was used as the loading control. (B) The qRT-PCR results showed that, compared to NC, the WT1 mRNA did not change significantly and FOXO1 mRNA decreased in the siATG5 group, while both WT1 and FOXO1 mRNA decreased significantly in the siBECN1 group. Data are presented as mean ± SD, n = 3, *P < 0.05, **P < 0.01 vs. NC. (C) After 0 h, 2 h, 4 h, and 6 h of CHX treatment to inhibit protein synthesis, Western blot was performed to detect the remaining amount of WT1 protein in KGN cells transfected with NC, siATG5, and siBECN1. GAPDH was used as the loading control. (D) The results of the quantitative analysis showed that the degradation rate of the WT1 protein in the siATG5 and siBECN1 groups was significantly slower compared to the NC. Data are presented as mean ± SD, n = 3. (E) Compared to control, there was a significant increase in WT1 protein level in KGN cells treated with CQ, but no significant change in FOXO1 protein. GAPDH was used as the loading control. (F and G) The degradation rate of WT1 protein in KGN cells pretreated with CQ was also delayed significantly. GAPDH was used as the loading control. Data are presented as mean ± SD, n = 3. It was noted that because the CHX chase assay was carried out after 48 h of siRNA infection or CQ treatment, the WT1 and SQSTM1 proteins had already accumulated in the siRNA or CQ group at 0 h (C and F). (H) Compared to control, the WT1 protein in the nucleus was elevated in KGN cells pretreated with CQ. LMNB1 and GAPDH were used as the loading controls for the nuclear and cytoplasmic proteins, respectively.
