Abstract
Background
A major issue with the current management of psoriasis is our inability to predict treatment response.
Objective
Our aim was to evaluate the ability to use baseline molecular expression profiling to assess treatment outcome for psoriatic patients.
Methods
We conducted a longitudinal study of 46 patients with chronic plaque psoriasis treated with anti-TNF agent etanercept, and molecular profiles were assessed in over 200 RNA-seq samples.
Results
We demonstrated correlation between clinical response and molecular changes during the course of the treatment, particularly for genes responding to IL-17A/TNF in keratinocytes. Intriguingly, baseline gene expressions in non-lesional, but not lesional skin, were the best marker of treatment response at week 12. We identified USP18, a known regulator of IFN responses, as positively correlated with PASI improvement (p=9.8×10−4) and demonstrate its role in regulating IFN/TNF responses in keratinocytes. Consistently, cytokine gene signatures enriched in baseline non-lesional skin expression profiles had strong correlations with PASI improvement. Using this information, we developed a statistical model for predicting PASI 75 (i.e. 75% of PASI improvement) at week 12, achieving AUC=0.75 and up to 80% accurate PASI75 prediction among the top predicted responders.
Conclusion
Our results illustrate feasibility of assessing drug response in psoriasis utilizing non-lesional skin and implicate involvement of IFN regulators in anti-TNF responses.
Keywords: psoriasis, etanercept, cytokine response, PASI, drug response prediction
Capsule summary
Enrichment of cytokine signatures in non-lesional psoriatic skin, prior to treatment, exhibits associations with future PASI improvement, and we illustrate the feasibility of integrating cytokine signals with transcriptomic data for drug response prediction in psoriasis.
INTRODUCTION
Psoriasis is an immune-mediated condition that affects the skin and joints of >100 million individuals worldwide. Patients with psoriasis have elevated tumor necrosis factor (TNF) in both lesional skin and blood (1, 2), and different agents inhibiting tumor necrosis factor (adalimumab, infliximab, golimumab, etanercept, certolizumab pegol) have been developed (3–7), and approved to treat moderate-to-severe psoriasis, making it one of the most commonly used biologic classes to treat psoriasis, and also recommended by expert for pregnancy/pediatric populations as well as patients with other comorbidities including psoriatic arthritis, cardiac disorders, and Crohn’s disease (8, 9).
Etanercept was amongst the first anti-TNF drugs approved to treat psoriasis. It is a fusion protein composed of two extracellular domains of the TNF receptor-2 fused to the human IgG1 Fc region. Etanercept binds and neutralizes soluble TNF (10) and lymphotoxin (11). Studies have illustrated its efficacy in reducing TNF expression in both non-lesional and lesional psoriatic skin (12). Furthermore, TNF inhibition has been shown to associate with reduced Th17 responses (13). Prior studies have demonstrated decreased Th17 immune response among clinical responders to etanercept (14). A double-blind study with 672 patients demonstrated that etanercept increases the proportion of patients achieving PASI75 (≥75% improvement of psoriasis severity index from baseline) within 12 weeks of treatment (3); however, similar to other anti-TNF agents, patient outcomes for etanercept treatment vary, with PASI75 responses ranging from 23% to 60% depending on the study (3, 15–18). Furthermore, the mechanisms involved in the difference in patient responses remain unclear and are incompletely explained by psoriasis susceptibility genes (19, 20). Providing assessment of drug responses prior to treatment may enhance treatment efficiency, limit the risk of unnecessary drug exposures, and reduce the economic burden for patients and society.
Precision medicine aims to identify which patients will respond best to a specific therapeutic approach for a disease (21–25). High-throughput advancements in the field of genomics have facilitated biomarker research and discovery endeavors, making precision medicine a more feasible approach to patient care. However, applications to complex skin conditions including psoriasis are still limited, despite available and extensive transcriptomic studies (20, 26–31). Here, we demonstrate that by applying statistical modeling using RNA-seq along with in vitro cytokine response genomic data to a cohort of etanercept-treated psoriatic patients, we can provide effective risk assessment for drug response. We demonstrate gene expression in healthy appearing non-lesional psoriatic skin at baseline is most predictive of therapeutic responses. Specifically, by using an integrative approach, we illustrate the enrichment of IFN and TNF signatures, and show how this prior information can be used in an analytical framework for predicting drug response.
METHODS
Patient cohort
Patients with moderate-to-severe chronic plaque psoriasis (sPGA ≥ 2) for longer than 6 months were enrolled for an open-label 50mg biweekly etanercept treatment for three months. Study protocols were approved by the University of Michigan IRB, and were carried out in accordance with Good Clinical Practice requirements and the Declaration of Helsinki. Informed consent was acquired from all participants. Patients were evaluated at baseline, and biopsy obtained under local anesthesia (lidocaine 1:10,000 epinephrine) from both non-lesional and lesional skin at baseline, and from lesional skin only at weeks 2, 6 and 12. Patients had full clinical assessment at 2, 6 and 12 weeks by a dermatologist and body-surface area, physician-global assessment and Psoriasis Area and Severity Index were recorded. The study was registered on clinicaltrials.gov (NCT01971346).
RNA-seq processing
We generated 50bp single-ended reads from 210 RNA-seq samples from 46 patients. For each sequence file, we conducted adapter trimming (32), and aligned reads to human genome hg19 (33). Only uniquely mapped reads were used for expression level quantification (34). We removed 1 patient due to drop out, and 2 patients that did not have baseline (week 0) biopsy samples, leaving 43 patients and 206 RNA-seq samples for the subsequent analysis. One patient’s last visit was conducted on week 15, and we grouped the results to the week 12 data for all other patients when conducting the analysis. We were able to detect 28,182 genes with on average ≥1 read/sample, and we applied DEseq2 for read normalization (35). Principal component analysis was conducted using all genes after applying inverse normalization for the DESeq2 normalized data.
Associating RNA-seq expression with clinical response
For each gene expression profile from baseline non-lesional or lesional skin samples, we correlated against change in PASI, BSA, and sPGA in each of the three follow-up visits. We adjusted the age, sex, and baseline BMI, and evaluated both percent (%) as well as absolute (i.e. delta) disease improvement referencing the week 0 values. False Discovery Rate (FDR) ≤10% was declared significant for association. Differential expression analyzes (i.e. comparing non-lesional vs lesional skin at baseline; comparing lesional skin at baseline versus subsequent visit) were conducted using DESeq2 negative binomial distribution, and FDR≤5% and |log2Fold Change| ≥1 were used as criteria to declare significant genes.
Comparison against cytokine signatures in keratinocytes
The procedures have been described previously (36). Briefly, we obtained 50 normal human keratinocytes from 50 different healthy adults. Keratinocytes were grown in 12 well plate in 154 CF medium (Thermo Fisher #M154CF500) with human keratinocyte growth supplement (Thermo Fisher #S0015). Keratinocytes were grown to confluency at which time the complete medium (with supplements) was replaced by basal 154 CF medium (without supplements). Cells were stimulated with cytokines (IL-4, IL-13, IFN-α, IFN-γ, TNF-α, IL-17A, R&D Systems) individually at 10 ng/ml concentration. After 8hrs cells were harvested and RNA was isolated using RNeasy Plus Mini kit (Qiagen # 74136). RNA was analyzed by RNA Nano Chips (Agilent Technologies) and sequenced (37). We extracted the top 1,000 genes with their baseline expression profiles showing the strongest correlations with future absolute PASI improvement in each of the three follow-up visits, and used hypergeometric test to compare against cytokine signatures to understand their molecular basis.
Predicting drug response
We assigned each patient from our cohort a TNF or IFN score using their baseline expression profiles of non-lesional skin: for each gene i induced by TNF/IFN in keratinocytes, we first computed the relative expression of that gene in patient p by referencing the median value across samples: rpi=gpi/median(gi), where g is the expression value after DESeq2 normalization; the TNF or IFN score for patient p was then defined as the upper quartile of the rp value across all genes induced by TNF or IFN, respectively(36, 38). To model the week 12 PASI response, we utilized the baseline non-lesional skin expression profiles for the genes induced by TNF/Type I IFN in keratinocytes. We used principal components to reduce the data dimension and applied logistic regression to model the drug responses at week 12 using the PASI75 criteria, using leave-one-out to ensure the model robustness (i.e. principal component analysis and regression modeling performed only on the training data). To assess the potential clinical implication, we measured the precision (proportion of true positives among the predicted PASI75) and recall (proportion of actual PASI 75 predicted) as functions of the proportion of top samples predicted from our model.
Immunohistochemistry
Formalin fixed, paraffin-embedded tissue slides obtained from patients with psoriasis and normal controls were heated for 30 min at 60°C, rehydrated, and epitope retrieved with Tris-EDTA, pH6. Slides were blocked, incubated with primary antibody UPS18 (1:100, LS-B1182–50, Lifespan Bioscience) overnight at 4°C. Slides were then washed with PBS and incubated with biotinylated secondary antibody (biotinylated goat anti-rabbit IgG Antibody, Vector Laboratories BA1000) for 30 min in room temperature, and then incubated with fluorochrome-conjugated streptavidin for 10 min in room temperature. Slides were prepared in mounting medium with 4’,6-diamidino-2-phenylindole (DAPI) (VECTASHIELD Antifade Mounting Medium with DAPI, H-1200, VECTOR). Images were acquired using Zeiss Axioskop 2 microscope and analyzed by SPOT software 5.1. Images presented are representative of at least three experiments.
RNAi -depletion, generation of USP18 overexpressing N/TERTs, RNA extraction, qRT-PCR
Upon reaching semi-confluence, the culture medium of keratinocytes was changed by Accell Delivery Media (B-005000, Dharmacon) with 1 μM Accell siRNA targeting USP18 (E-004236–00-0005, Dharmacon). After 48h, cells were stimulated with IFN-α (10ng/ml, R&D Systems; I4276), TNF (10ng/ml, R&D Systems: 210-TA), IL-17A (20ng/ml, R&D Systems; 7955-IL), IFN-γ (10ng/ml, R&D Systems: 285-IF) for another 24h. RNA was isolated from cells using Qiagen RNeasy plus kit (74136). QRT-PCR was performed on a 7900HT Fast Real-time PCR system (Applied Biosystems) with TaqMan Universal PCR Master Mix (ThermoFisher 4304437). N/TERT keratinocytes stably overexpressing USP18 were generated using 4D-Nucleofector X Unit (Lonza Cologne, Germany). Cells were prepared using standard protocol for Normal Human Epidermal Keratinocyte X Unit kit (4D Nucleofector Solution, supplement and 100μL single nucleocuvette) obtained from Lonza. For each electroporation, 5μg pCMV6-AC-GFP USP18 plasmid (Origene, Rockville, Maryland, USA) was used. Unit X program used was DS-138 for stable keratinocytes. Following transfection, keratinocytes were grown in a 12-well plate using fully supplemented Keratinocyte-SFM medium, penicillin streptomycin and 500μg/mL G418 (Geneticin by Thermo Fisher Scientific (Waltham, Massachusetts, USA)) for selection followed by expansion for approximately 30 days. USP18-GFP overexpression was validated using qRT-PCR and western blotting. Primers (ThermoFisher Scientific) used in this study were: USP18, Hs00276441_m1; IL36G, Hs00219742_m1; DEFB4, Hs00175474_m1; MX1, Hs00895608_m1; OASL, Hs00984387_m1; IRF7, Hs01014809_g1; IFNK, Hs00737883_m1.
RESULTS
RNA-seq to profile transcriptomic changes in patients initiated on etanercept therapy
We enrolled 46 psoriatic patients for this study (Figure 1a). Each patient was treated with etanercept, 50mg twice weekly. Prior to initiation (baseline/week 0), demographic (age, sex) and other clinical information including psoriasis area and severity index (PASI), body surface area (BSA), and static physician’s global assessment (sPGA) were collected. Biopsies were performed at baseline on non-lesional and lesional psoriatic skin for transcriptome profiling using RNA-seq, and additional lesional samples were obtained at time of follow-up on week 2, week 6, and week 12, along with clinical assessment. Follow-up data on 42 patients was obtained (i.e. with transcriptomic data from both baseline and at least one of the follow-up visits), and a total of 36 patients completed the study with RNA-seq and clinical data available at both baseline and week 12 visit. A total of 210 RNA-seq experiments were performed.
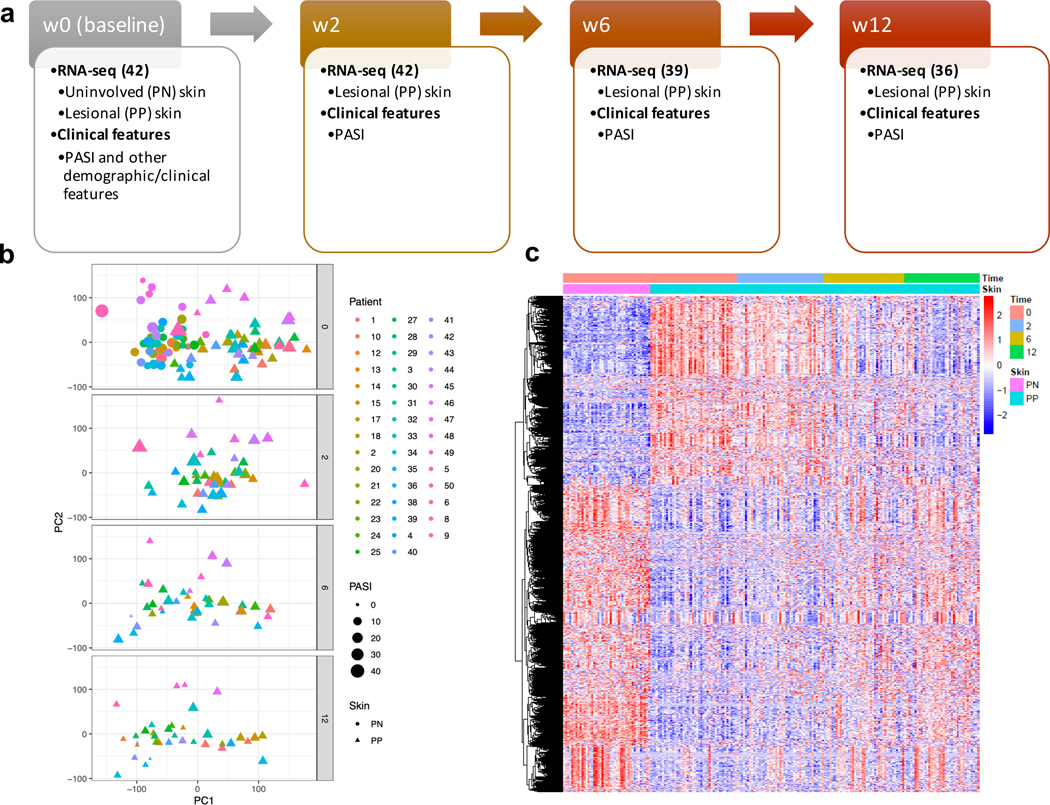
Figure 1. Transcriptome of the longitudinal cohort.
a) Design of the study; b) the top two principal components computed using transcriptomic data for all the RNA-seq samples; c) heatmap illustrating the change in expression profiles across the treatment time course. Note 46 patients were recruited in our cohort, and 42 patients have RNA-seq data from both baseline and at least one of the follow-up visits. The union set of genes that are differentially expressed between baseline lesional skin versus follow-up lesional skin samples was used to construct this heatmap.
By using principal component analysis (PCA), we tracked transcriptomic changes over the treatment course (Figure 1b). As expected, PCA separated non-lesional and lesional skin at baseline, but during the treatment period, the largest principal component (i.e. PC1) of lesional skin changed over time such that by week 12 more lesional skin samples overlapped with that of non-lesional skin at baseline, while the PC1 of some lesional skin samples still remained at baseline level, despite improvement in PASI by week 12. Notably, the former group tended to have a lower PASI score than the latter group at week 12 (lower panel in Figure 1b). We further investigated this at the gene expression level and demonstrated how the expression profiles changed during the 12-week treatment (Figure 1c). Similar to the PCA results, there was a sharp contrast between non-lesional and lesional psoriatic skin at baseline, but the contrast became less pronounced as treatment proceeded. Indeed, when comparing gene expression in baseline lesional skin with that from follow-up visits, we observed a gradual increase in the number of differentially expressed transcripts (Supplementary Table 1), and restoration towards gene expression levels in non-lesional skin at baseline (Supplementary Figure 1). Nevertheless, heterogeneity was observed among patients’ transcriptomic responses at week 12, concordant with the clinical variations (3, 14).
Expression profiles at baseline non-lesional psoriatic skin are associated with PASI improvement
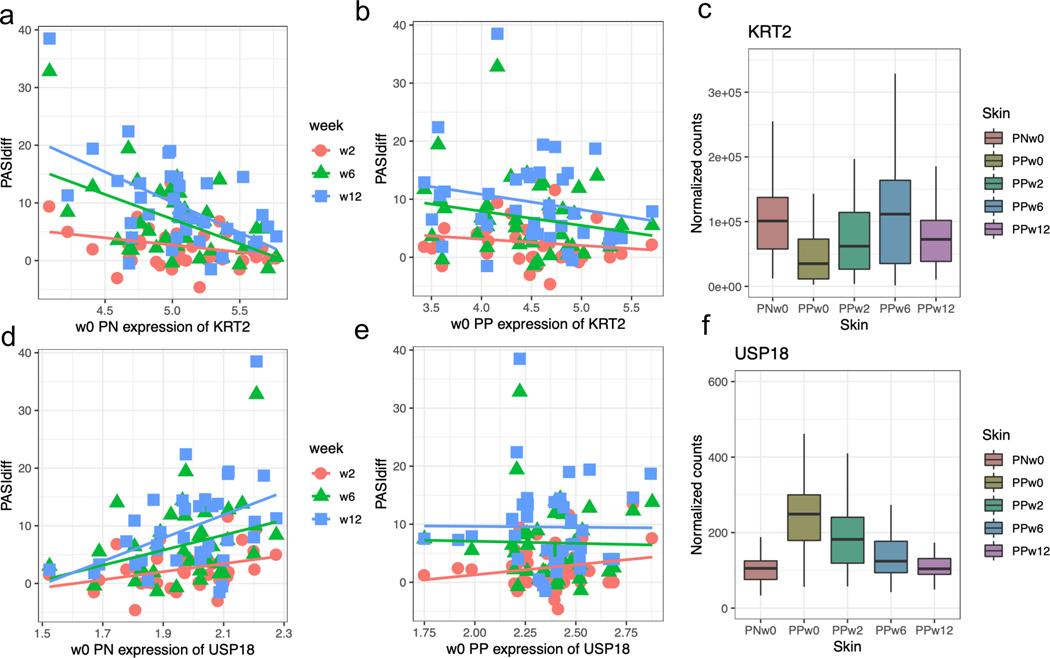
To examine the associations between baseline expression profiles and clinical presentation, expression level (inverse normalized to ensure robustness) from non-lesional or lesional skin of each gene at week 0 was correlated with the change in PASI, BSA, and sPGA in each of the three follow-up visits. We evaluated both percent (%) as well as absolute (i.e. delta) disease improvement referencing the week 0 values. Surprisingly, we could only identify significant associations for gene profiles in baseline non-lesional skin, rather than lesional skin. When using percent change as measure there were 198 genes with their week 0 expression profiles significantly (False Discovery Rate, FDR≤10%) associated with week 12 PASI improvement; when using absolute change as measure, there were 192 and 391 genes with their baseline expression profiles significantly associated with week 6 and week 12 PASI improvement, respectively. We were not able to reveal any significant results for associations against BSA and sPGA change. Among the significant genes with their baseline expressions associating with PASI improvement at week 12, 105 overlapped between the percent and absolute measures. USP18, an ubiquitin specific peptidase, and KRT2, a type I cytokeratin, were two prominent examples with expression in non-lesional skin at baseline showing significant association with PASI improvement in follow-up visit (Figure 2): USP18 was significantly up-regulated in lesional skin at baseline (fold change, FC=2.5; p=1×10−26), and its expression in non-lesional skin at baseline was positively correlated with absolute PASI improvement at week 12 (p=9.8×10−4; 20% increase from mean expression level had on average 2.3 PASI improvement after adjusting for age, sex, and BMI); KRT2 was significantly down-regulated in lesional skin at baseline (FC=0.32; p=1.3×10−6) and its expression in non-lesional skin at week 0 was inversely correlated with both the absolute (p=1.4×10−5; 20% decrease from mean expression level has on average 0.99 PASI improvement after adjusting for age, sex, and BMI) and percent (p=5.4×10−4) PASI improvement at week 12. Although their associations with PASI improvement at week 2 and week 6 were not significant, the direction of correlations was consistent (Figure 2a, d); interestingly, these associations were not observed when using baseline expression levels in lesional skin (Figure 2b, e). Furthermore, the expressions of these two genes in lesional skin gradually “normalized” towards the expression levels in non-lesional skin with treatment (Figure 2c, f). These results indicate that both USP18 and KRT2 are dysregulated in psoriatic skin but can be “restored” to non-lesional skin levels by etanercept treatment, and their expressions in non-lesional skin prior to treatment correlate with PASI improvement.
Figure 2. The associations between PASI improvement and baseline expression.
The PASI improvement (y-axis) was plotted against the baseline expressions of KRT2 (a, b) and USP18 (d, e) in non-lesional and lesional skin; the boxplots for normalized expression levels of KRT2 (c) and USP18 (f) in different skin types and time points during the treatment course are shown in c and f.
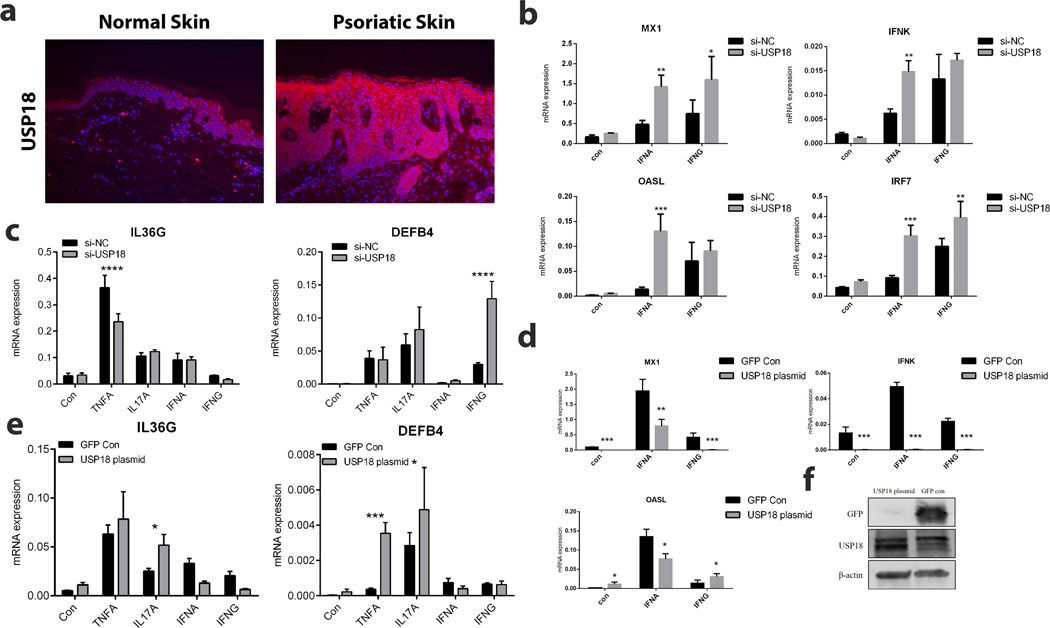
To understand the role of USP18 in TNF responses, we used an independent transcriptomic cohort for psoriasis (36) to validate the up-regulation of USP18 in lesional skin (Supplementary Figure 2); interestingly, we also observed that its expression was marginally up-regulated in non-lesional skin when compared with healthy control skin (FC=1.5; p=1.2×10−2). USP18 was also positively correlated with expression of other psoriasis cytokines (IL23A and IL36G) in lesional skin (p=6.3×10−5 and p=8.3×10−7, respectively). Immunostaining (Figure 3a) confirmed expression of USP18 in the epidermal layer in both non-lesional and lesional psoriatic skin. Knockdown of USP18 using siRNA (Supplementary Figure 3) was done in keratinocytes to assess the impact on TNF/IFN responses. Effective depletion of USP18 affected both TNF and IFN response (Figure 3b, c), for instance, IL36G mRNA expression was suppressed upon TNF stimulation but DEFB4 was induced upon IFN-γ stimulation). Similarly, USP18 knock-down promoted higher expression of IFN-responding genes, including MX1, IFNK, OASL, and IRF7. Conversely, USP18 overexpression decreased type I and type II IFN responses (Figure 3d) but enhanced IL-17A induced effect on IL36G and TNF-induced effect on DEFB4 (Figure 3e). These results suggest that USP18 shifts the balance between IFN and TNF responses in keratinocytes. As these two signals have been thought to have counter-regulatory roles in psoriasis (39), lower USP18 might promote higher IFN response and thus lower TNF dependence, and vice versa, agreeing with the positive correlation with PASI improvement we observed during the course of etanercept treatment.
Figure 3. USP18 as a modulator for IFN/TNF response.
a) Immunostaining for USP18 in normal and psoriatic skin; b) effect of depleting USP18 on type I and type II stimulations (x-axis); c) effect of depleting USP18 on expression of IL36G and DEFB4 upon TNF, IL-17A, IFN-α and IFN-γ stimulations (x-axis); d) effect of USP18 overexpression on type I and type II IFN responses; e) effect of USP18 overexpression on expression of IL36G and DEFB4 upon TNF, IL-17A, IFN-α and IFN-γ stimulations; f) Western blot confirming increased USP18 protein levels in USP18 plasmid transfected cells. Data shown with STDV and are representative of 3 biologic replicates. * p<0.05, **p<0.01, *** p<0.001.
Integrative approach to provide biological and clinical implications using transcriptomic data from non-lesional psoriatic skin
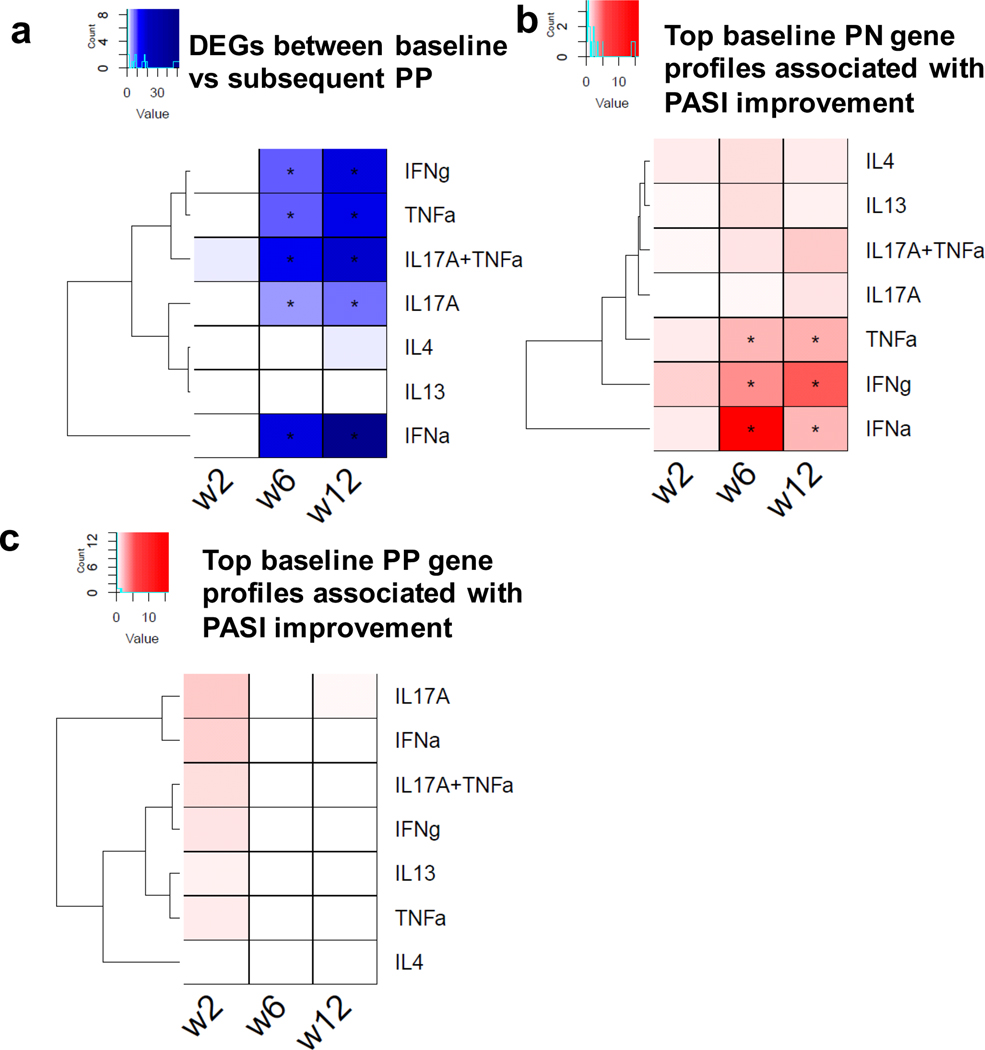
To provide biological and clinical implications for etanercept treatment in psoriasis, we utilized cytokine signatures in keratinocytes obtained from independent RNA-seq transcriptomic data (36, 38). Among the genes showing the strongest differential expression in lesional skin between baseline and subsequent follow-up visits, we observed significant (FDR≤5%) enrichment of IFN, TNF, and IL-17 response genes (Figure 4a), concordant with the drug’s TNF inhibitory effect and its associated negative regulation on Th17 response (14, 15). Next, we extracted the top 1,000 genes with their baseline expression profiles showing the strongest correlations with absolute PASI improvement in each of the three follow-up visits, and compared them against type I IFN, TNF, and IL-17A keratinocyte cytokine signatures to understand their molecular basis. Strikingly, we found significant enrichment for TNF and Type I IFN signatures for week 0 non-lesional skin gene expression profiles which have the strongest correlations with week 6 and week 12 absolute PASI improvement (Figure 4b). Notably, there was no significant enrichment when using week 0 gene expression profiles in lesional skin (Figure 4c). When using percentage PASI improvement as a measure, we observed the same Type I IFN signature enrichment (week 6 for IFN-α and week 12 for IFN-γ), and we also revealed enrichment for IL-17A responses at week 12. It is notable that both USP18 (Supplementary Figures 3 and 4) and KRT2 were differentially expressed upon TNF stimulation in keratinocytes, and USP18 was also upregulated by IFNs.
Figure 4. Enrichment of different cytokine signatures in different association comparisons.
a) Enrichment of the signatures among genes showing strongest differential expression in lesional skin between baseline versus follow-up; b,c) enrichment of the signatures among expression profiles at baseline in non-lesional psoriatic skin (b) or lesional skin (c) having strongest association with follow-up PASI improvement.
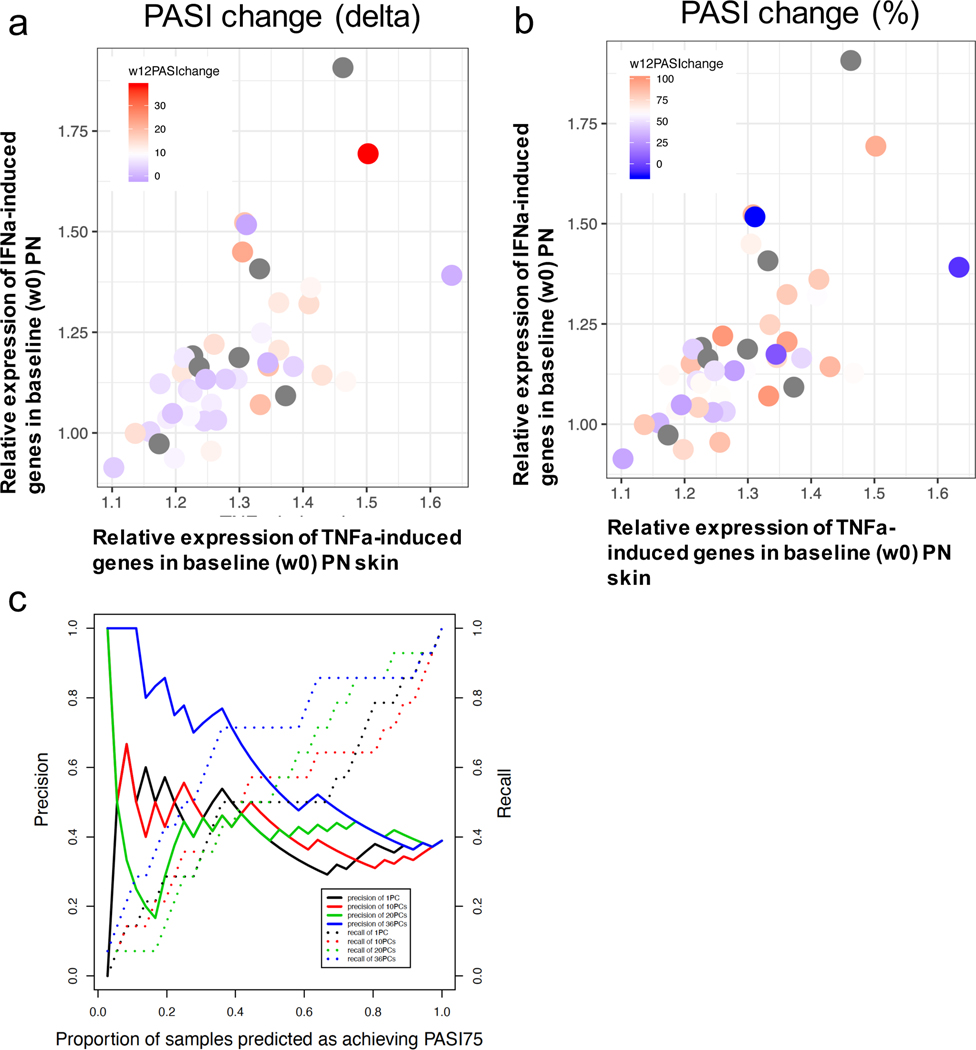
We assigned each patient from our cohort a TNF or IFN score (see Methods), summarizing the respective cytokine signature loading for the non-lesional skin of that patient at baseline (Figure 5a, b). Among the patients showing higher IFN/TNF scores, we observed a larger proportion for PASI improvement, in terms of both absolute or percentage measures. By using keratinocyte cytokine response signatures as prior information, we computed, for each patient, the principal components of the baseline non-lesional skin expression levels for >2,900 genes that were induced in TNF, IFN-α, or IFN-γ stimulations. We applied logistic regression to these components to model drug responses at week 12 using the PASI 75 criteria, using the leave-one-out cross validation to ensure robustness upon model evaluation, and were able to obtain up to 0.75 in area under the receiver operating characteristic (AUC). To further assess the potential clinical implications, we measured the precision (proportion of true positives among the predicted PASI75) and recall (proportion of actual PASI75 predicted) among the top samples predicted from our model (Figure 5c). For the top 20% of samples exhibiting the top PASI 75 prediction, we achieved up to 80% accuracy, representing the ability to use baseline non-lesional psoriatic skin transcriptome to identify patients that benefit most from etanercept treatment.
Figure 5. Assessment of PASI response in week 12 using baseline non-lesional skin expression profiles.
a,b) Each patient’s TNF score at baseline non-lesional psoriatic skin (x-axis) was plotted against the patient’s IFN score, and the color scheme (grey color represents no follow-up data) represents the PASI improvement by week 12 using either absolute (a) or percent (b) measure; c) the precision (solid line) and recall (dotted line) were plotted against the top proportion of samples predicted to achieve PASI75 by week 12.
DISCUSSION
The use of targeted biologic drugs has revolutionized the treatment of psoriasis. The anti-TNF class includes different agents and has been in use for psoriasis for close to two decades (3). However, biologics have different effectiveness (40), and clinical responses may vary widely across patients (41), despite otherwise highly similar or identical disease. The reasons for this variability have remained unclear until now, and are incompletely explained by the contribution of genetic susceptibility variants (42, 43). Together with the fact that biologic treatments pose a huge economic burden on both patients and society (44, 45), information regarding how each patient’s characteristics associate with treatment outcomes is urgently needed.
Previous studies using gene expression microarray data have demonstrated that response to the anti-TNF agent etanercept is accompanied by suppression of Th17 and TNF signaling pathways (14, 46). However, as IL-17 responses are prominent in practically all patients with psoriasis (47), this observation has not led to ways to predict treatment responses. In this study, we used RNA-seq to profile the skin prior to and during the course of etanercept treatment of psoriasis. Our results unexpectedly demonstrate that baseline expression data from non-lesional skin, rather than lesional psoriatic skin, is a better predictor of clinical response to etanercept.
It has been established in multiple studies that non-lesional skin from psoriatic patients is different than healthy control skin. Noted differences include an increased level of epidermal proliferation (48); lower levels of the epidermal barrier proteins, e.g. filaggrin and loricrin (49), changes in innate immune response and lipid metabolism genes, e.g. S100A7 and ELOVL3 (28), and an abnormal epidermal barrier recovery (50). The reason for these changes in non-lesional psoriatic skin is unclear but speculated to reflect a systemic response from increased levels of circulating pro-inflammatory mediators (51), genetic predisposition (52, 53), or a combination of both. This suggests that while expression profiles from non-lesional skin may provide predictive power in PASI improvement, expression profiles of lesional skin in turn provide richer information for understanding pathogenic mechanisms of psoriasis in general, as has been demonstrated in multiple studies (26, 27). Thus, in the chronic inflammatory state of lesional psoriatic skin, where multiple different cytokines act locally, constituting a heterogeneous environment to study context-specific inflammatory response (54), non-lesional skin likely represents a state with refined resolution to decipher subtle variations of individual and likely systemic pro-inflammatory state, thereby enhancing predictions of therapeutic response.
We were able to associate non-lesional expression profiles at baseline with both percent and absolute PASI improvement and using PASI75 to categorize responders versus non-responders. While percent improvement in PASI is a more conventional metric in psoriasis studies, in some scenario it does not distinguish well between patients with different disease severity at baseline. For example, patient with a drop from 40 to 10 in PASI score has the same PASI 75 response as someone dropping from 10 down to 2.5. As the inflammatory load would be expected to be much higher in the first compared to the second patient, using an absolute change in PASI (delta PASI of 30 in the first patient vs. 7.5 in the second) can potentially provide a greater likelihood of detecting a systemic inflammatory signal in non-lesional skin. To determine if this approach leads to a bias towards disease severity, we correlated the PASI75 with baseline PASI score, and did not identify significant changes, suggesting that our observation was not dependent upon disease severity.
Identification of USP18 as a predictor of therapeutic response to etanercept is consistent with the cross-regulation that is known to occur between type I IFNs and TNF (55). Notably, TNF regulates type I IFN production by blocking development of plasmacytoid dendritic cells (pDCs), as well as inhibiting its secretion; in contrast, blocking TNF sustains type I IFN release from pDCs (55). An example of this is so called “paradoxical psoriasis” that arises in the setting of anti-TNF treatment. It is a relatively uncommon reaction occurring in approximately 2–5% of patients with psoriasis and is characterized by accumulation of pDCs and overexpression of type I IFNs (39). Our data suggests another source of cross-regulation between these two cytokines at the level of the keratinocyte, involving the ubiquitin specific peptidase; USP18. Consistent with our findings, USP18 has been implicated as a negative regulator of IFN signaling (56), and a promoter of TNF responses (57).
Given the cross-regulation of TNF and IFN responses outlined above it is worth noting that we observe both of these pro-inflammatory signals in non-lesional skin (Figure 4). However, these are averaged across our cohort of >36 patients and therefore do not fully reflect the balance on an individual level at baseline between these two signals. Furthermore, while we see suppression of IL-17 responses with effective anti-TNF treatment (13), it is noteworthy that the strength of the IL-17 responses at baseline in lesional or non-lesional skin was not as predictive of etanercept treatment response. IL-17 is a critical component of the inflammatory network in psoriasis, as outlined by both genetic and clinical studies (58, 59). Notably, IL-17 responses synergize with TNF (60), but, interestingly, it can also synergize with IFNs, particularly IFN-γ (61, 62), suggesting that both of these inflammatory circuits can work with IL-17 to maintain disease activity. It is likely that predictors for treatment responses to anti-IL-17 and anti-IL23 agents would be different than that for etanercept. Anti-TNF treatment is prescribed to ~20% of psoriatic patients, particularly those that have concomitant psoriatic arthritis, and in a recent study on prescription patterns associated with biologic therapies for psoriasis from a U.S Medical Records database (63), etanercept was the second most prescribed biologic for psoriasis (second only to adalimumab). As etanercept has been shown to have decreased risk of infection compared to other anti TNF agents (64), the benefit from being able to predict response to a lesser effective biologic, but one with decreased risk of broader immunosuppression and infectious complications, is clear. Future studies are needed evaluate if the biological predictors identified in this study can be used to provide drug response assessment for other anti-TNF agents.
In summary, our data illustrates that transcriptomic data can provide distinguishing power for individuals carrying extreme IFN/TNF burden in non-lesional psoriatic skin. Our findings are important as they show the feasibility of using transcriptomic approaches for drug prediction in psoriasis.
Supplementary Material
Key messages.
We illustrated USP18 association between baseline non-lesional psoriatic skin expression with PASI improvement, and show USP18 has inhibitory effect on IFN response, while facilitating TNF responses.
By using cytokine signatures from keratinocytes, we assessed drug response, obtaining up to 0.75 in area under the receiver operating characteristic (AUC).
Acknowledgements
This work was supported by Amgen. LCT, MKS, JEG were supported by the Babcock Endowment Fund. We also received supported from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS): R01-AR060802, P30-AR075043 (JEG), and AR072129 (LCT), the National Institute of Allergy and Infectious Diseases (NIAID) under Award Number R01-AR069071 (JEG), the A. Alfred Taubman Medical Research Institute Kenneth and Frances Eisenberg Emerging Scholar Award (JEG), Taubman Medical Institute (JEG, JMK, LCT), the Dermatology Foundation (LCT), the Arthritis National Research Foundation (LCT), and the National Psoriasis Foundation Psoriasis Prevention Initiative (LCT, JEG, EM, JMK).
This work was supported by Amgen. JEG received research grants from AbbVie, AnaptysBio, SunPharma, Genentech, Pfizer, Novartis, Celgene, Almirall, and Eli Lilly, and serves as advisory board in Novartis, AbbVie, Eli Lilly, MiRagen, BMS/Celgene, AstraZeneca, and Almirall. J.M.K serves on an advisory board for AstraZeneca.
Abbreviations
- AUC
area under the receiver operating characteristic
- BSA
body surface area
- FDR
false discovery rate
- PASI
psoriasis area and severity index
- PCA
principal component analysis
- sPGA
static physician’s global assessment
Footnotes
Conflict of interest
The other authors report no conflicts of interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Arican O, Aral M, Sasmaz S, and Ciragil P. Serum levels of TNF-alpha, IFN-gamma, IL-6, IL-8, IL-12, IL-17, and IL-18 in patients with active psoriasis and correlation with disease severity. Mediators Inflamm. 2005;2005(5):273–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johansen C, Funding AT, Otkjaer K, Kragballe K, Jensen UB, Madsen M, et al. Protein expression of TNF-alpha in psoriatic skin is regulated at a posttranscriptional level by MAPK-activated protein kinase 2. J Immunol. 2006;176(3):1431–8. [DOI] [PubMed] [Google Scholar]
- 3.Leonardi CL, Powers JL, Matheson RT, Goffe BS, Zitnik R, Wang A, et al. Etanercept as monotherapy in patients with psoriasis. N Engl J Med. 2003;349(21):2014–22. [DOI] [PubMed] [Google Scholar]
- 4.Gordon KB, Langley RG, Leonardi C, Toth D, Menter MA, Kang S, et al. Clinical response to adalimumab treatment in patients with moderate to severe psoriasis: double-blind, randomized controlled trial and open-label extension study. Journal of the American Academy of Dermatology. 2006;55(4):598–606. [DOI] [PubMed] [Google Scholar]
- 5.Chaudhari U, Romano P, Mulcahy LD, Dooley LT, Baker DG, and Gottlieb AB. Efficacy and safety of infliximab monotherapy for plaque-type psoriasis: a randomised trial. Lancet. 2001;357(9271):1842–7. [DOI] [PubMed] [Google Scholar]
- 6.Kavanaugh A, McInnes I, Mease P, Krueger GG, Gladman D, Gomez-Reino J, et al. Golimumab, a new human tumor necrosis factor alpha antibody, administered every four weeks as a subcutaneous injection in psoriatic arthritis: Twenty-four-week efficacy and safety results of a randomized, placebo-controlled study. Arthritis Rheum. 2009;60(4):976–86. [DOI] [PubMed] [Google Scholar]
- 7.Blauvelt A, Reich K, Lebwohl M, Burge D, Arendt C, Peterson L, et al. Certolizumab pegol for the treatment of patients with moderate-to-severe chronic plaque psoriasis: pooled analysis of week 16 data from three randomized controlled trials. J Eur Acad Dermatol Venereol. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaushik SB, and Lebwohl MG. Psoriasis: Which therapy for which patient: Psoriasis comorbidities and preferred systemic agents. Journal of the American Academy of Dermatology. 2019;80(1):27–40. [DOI] [PubMed] [Google Scholar]
- 9.Kaushik SB, and Lebwohl MG. Psoriasis: Which therapy for which patient: Focus on special populations and chronic infections. Journal of the American Academy of Dermatology. 2019;80(1):43–53. [DOI] [PubMed] [Google Scholar]
- 10.Gisondi P, and Girolomoni G. Biologic therapies in psoriasis: a new therapeutic approach. Autoimmun Rev. 2007;6(8):515–9. [DOI] [PubMed] [Google Scholar]
- 11.Mohler KM, Torrance DS, Smith CA, Goodwin RG, Stremler KE, Fung VP, et al. Soluble tumor necrosis factor (TNF) receptors are effective therapeutic agents in lethal endotoxemia and function simultaneously as both TNF carriers and TNF antagonists. J Immunol. 1993;151(3):1548–61. [PubMed] [Google Scholar]
- 12.Caldarola G, De Simone C, Carbone A, Tulli A, Amerio P, and Feliciani C. TNFalpha and its receptors in psoriatic skin, before and after treatment with etanercept. Int J Immunopathol Pharmacol. 2009;22(4):961–6. [DOI] [PubMed] [Google Scholar]
- 13.Zaba LC, Cardinale I, Gilleaudeau P, Sullivan-Whalen M, Suarez-Farinas M, Fuentes-Duculan J, et al. Amelioration of epidermal hyperplasia by TNF inhibition is associated with reduced Th17 responses. J Exp Med. 2007;204(13):3183–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zaba LC, Suarez-Farinas M, Fuentes-Duculan J, Nograles KE, Guttman-Yassky E, Cardinale I, et al. Effective treatment of psoriasis with etanercept is linked to suppression of IL-17 signaling, not immediate response TNF genes. J Allergy Clin Immunol. 2009;124(5):1022–10 e1–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krueger GG, Elewski B, Papp K, Wang A, Zitnik R, and Jahreis A. Patients with psoriasis respond to continuous open-label etanercept treatment after initial incomplete response in a randomized, placebo-controlled trial. Journal of the American Academy of Dermatology. 2006;54(3 Suppl 2):S112–9. [DOI] [PubMed] [Google Scholar]
- 16.Tyring S, Gordon KB, Poulin Y, Langley RG, Gottlieb AB, Dunn M, et al. Long-term safety and efficacy of 50 mg of etanercept twice weekly in patients with psoriasis. Arch Dermatol. 2007;143(6):719–26. [DOI] [PubMed] [Google Scholar]
- 17.Paller AS, Siegfried EC, Eichenfield LF, Pariser D, Langley RG, Creamer K, et al. Long-term etanercept in pediatric patients with plaque psoriasis. Journal of the American Academy of Dermatology. 2010;63(5):762–8. [DOI] [PubMed] [Google Scholar]
- 18.Kivelevitch D, Mansouri B, and Menter A. Long term efficacy and safety of etanercept in the treatment of psoriasis and psoriatic arthritis. Biologics. 2014;8:169–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foulkes AC, and Brown SJ. Genetic prediction of treatment response in psoriasis is still a work in progress. Br J Dermatol. 2017;177(2):344–5. [DOI] [PubMed] [Google Scholar]
- 20.Tsoi LC, Yang J, Liang Y, Sarkar MK, Xing X, Beamer MA, et al. Transcriptional determinants of individualized inflammatory responses at anatomically separate sites. J Allergy Clin Immunol. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aziz MA, Yousef Z, Saleh AM, Mohammad S, and Al Knawy B. Towards personalized medicine of colorectal cancer. Crit Rev Oncol Hematol. 2017;118:70–8. [DOI] [PubMed] [Google Scholar]
- 22.Chan CWH, Law BMH, So WKW, Chow KM, and Waye MMY. Novel Strategies on Personalized Medicine for Breast Cancer Treatment: An Update. Int J Mol Sci. 2017;18(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flamant M, and Roblin X. Inflammatory bowel disease: towards a personalized medicine. Therap Adv Gastroenterol. 2018;11:1756283X17745029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horton I, Lin Y, Reed G, Wiepert M, and Hart S. Empowering Mayo Clinic Individualized Medicine with Genomic Data Warehousing. J Pers Med. 2017;7(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tavakolpour S. Towards personalized medicine for patients with autoimmune diseases: Opportunities and challenges. Immunol Lett. 2017;190:130–8. [DOI] [PubMed] [Google Scholar]
- 26.Li B, Tsoi LC, Swindell WR, Gudjonsson JE, Tejasvi T, Johnston A, et al. Transcriptome analysis of psoriasis in a large case-control sample: RNA-seq provides insights into disease mechanisms. J Invest Dermatol. 2014;134(7):1828–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsoi LC, Iyer MK, Stuart PE, Swindell WR, Gudjonsson JE, Tejasvi T, et al. Analysis of long non-coding RNAs highlights tissue-specific expression patterns and epigenetic profiles in normal and psoriatic skin. Genome Biol. 2015;16:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gudjonsson JE, Ding J, Li X, Nair RP, Tejasvi T, Qin ZS, et al. Global gene expression analysis reveals evidence for decreased lipid biosynthesis and increased innate immunity in uninvolved psoriatic skin. J Invest Dermatol. 2009;129(12):2795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gudjonsson JE, Ding J, Johnston A, Tejasvi T, Guzman AM, Nair RP, et al. Assessment of the psoriatic transcriptome in a large sample: additional regulated genes and comparisons with in vitro models. J Invest Dermatol. 2010;130(7):1829–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merleev AA, Marusina AI, Ma C, Elder JT, Tsoi LC, Raychaudhuri SP, et al. Meta-analysis of RNA sequencing datasets reveals an association between TRAJ23, psoriasis, and IL-17A. JCI Insight. 2018;3(13). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le ST, Merleev AA, Luxardi G, Shimoda M, Adamopoulos IE, Tsoi LC, et al. 2D Visualization of the Psoriasis Transcriptome Fails to Support the Existence of DualSecreting IL-17A/IL-22 Th17 T Cells. Front Immunol. 2019;10:589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bolger AM, Lohse M, and Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anders S, Pyl PT, and Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Love MI, Huber W, and Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsoi LC, Rodriguez E, Degenhardt F, Baurecht H, Wehkamp U, Volks N, et al. Atopic dermatitis is an IL-13 dominant disease with greater molecular heterogeneity compared to psoriasis. J Invest Dermatol. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarkar MK, Hile GA, Tsoi LC, Xing X, Liu J, Liang Y, et al. Photosensitivity and type I IFN responses in cutaneous lupus are driven by epidermal-derived interferon kappa. Ann Rheum Dis. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsoi LC, Rodriguez E, Stolzl D, Wehkamp U, Sun J, Gerdes S, et al. Progression of acute-to-chronic atopic dermatitis is associated with quantitative rather than qualitative changes in cytokine responses. J Allergy Clin Immunol. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conrad C, Di Domizio J, Mylonas A, Belkhodja C, Demaria O, Navarini AA, et al. TNF blockade induces a dysregulated type I interferon response without autoimmunity in paradoxical psoriasis. Nat Commun. 2018;9(1):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loos AM, Liu S, Segel C, Ollendorf DA, Pearson SD, and Linder JA. Comparative effectiveness of targeted immunomodulators for the treatment of moderate-to-severe plaque psoriasis: A systematic review and network meta-analysis. Journal of the American Academy of Dermatology. 2018;79(1):135–44 e7. [DOI] [PubMed] [Google Scholar]
- 41.Vide J, and Magina S. Moderate to severe psoriasis treatment challenges through the era of biological drugs. An Bras Dermatol. 2017;92(5):668–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tejasvi T, Stuart PE, Chandran V, Voorhees JJ, Gladman DD, Rahman P, et al. TNFAIP3 gene polymorphisms are associated with response to TNF blockade in psoriasis. J Invest Dermatol. 2012;132(3 Pt 1):593–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swindell WR, Xing X, Stuart PE, Chen CS, Aphale A, Nair RP, et al. Heterogeneity of inflammatory and cytokine networks in chronic plaque psoriasis. PloS one. 2012;7(3):e34594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu JJ, Feldman SR, Rastogi S, Menges B, Lingohr-Smith M, and Lin J. Comparison of the cost-effectiveness of biologic drugs used for moderate-to-severe psoriasis treatment in the United States. J Dermatolog Treat. 2018:1–6. [DOI] [PubMed] [Google Scholar]
- 45.Klijn SL, van den Reek J, van de Wetering G, van der Kolk A, de Jong E, and Kievit W. Biologic treatment sequences for plaque psoriasis: a cost-utility analysis based on 10 years of Dutch real-world evidence from BioCAPTURE. Br J Dermatol. 2018;178(5):1181–9. [DOI] [PubMed] [Google Scholar]
- 46.Johnston A, Guzman AM, Swindell WR, Wang F, Kang S, and Gudjonsson JE. Early tissue responses in psoriasis to the antitumour necrosis factor-alpha biologic etanercept suggest reduced interleukin-17 receptor expression and signalling. Br J Dermatol. 2014;171(1):97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swindell WR, Sarkar MK, Liang Y, Xing X, and Gudjonsson JE. Cross-Disease Transcriptomics: Unique IL-17A Signaling in Psoriasis Lesions and an Autoimmune PBMC Signature. J Invest Dermatol. 2016;136(9):1820–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krueger GG, Chambers DA, and Shelby J. Involved and uninvolved skin from psoriatic subjects: are they equally diseased? Assessment by skin transplanted to congenitally athymic (nude) mice. J Clin Invest. 1981;68(6):1548–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim BE, Howell MD, Guttman-Yassky E, Gilleaudeau PM, Cardinale IR, Boguniewicz M, et al. TNF-alpha downregulates filaggrin and loricrin through c-Jun N-terminal kinase: role for TNF-alpha antagonists to improve skin barrier. J Invest Dermatol. 2011;131(6):1272–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ye L, Lv C, Man G, Song S, Elias PM, and Man MQ. Abnormal epidermal barrier recovery in uninvolved skin supports the notion of an epidermal pathogenesis of psoriasis. J Invest Dermatol. 2014;134(11):2843–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dowlatshahi EA, van der Voort EA, Arends LR, and Nijsten T. Markers of systemic inflammation in psoriasis: a systematic review and meta-analysis. Br J Dermatol. 2013;169(2):266–82. [DOI] [PubMed] [Google Scholar]
- 52.Tsoi LC, Spain SL, Knight J, Ellinghaus E, Stuart PE, Capon F, et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nature genetics. 2012;44(12):1341–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsoi LC, Stuart PE, Tian C, Gudjonsson JE, Das S, Zawistowski M, et al. Large scale meta-analysis characterizes genetic architecture for common psoriasis associated variants. Nat Commun. 2017;8:15382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim-Hellmuth S, Bechheim M, Putz B, Mohammadi P, Nedelec Y, Giangreco N, et al. Genetic regulatory effects modified by immune activation contribute to autoimmune disease associations. Nat Commun. 2017;8(1):266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Palucka AK, Blanck JP, Bennett L, Pascual V, and Banchereau J. Cross-regulation of TNF and IFN-alpha in autoimmune diseases. Proc Natl Acad Sci U S A. 2005;102(9):3372–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arimoto KI, Lochte S, Stoner SA, Burkart C, Zhang Y, Miyauchi S, et al. STAT2 is an essential adaptor in USP18-mediated suppression of type I interferon signaling. Nat Struct Mol Biol. 2017;24(3):279–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shaabani N, Honke N, Nguyen N, Huang Z, Arimoto KI, Lazar D, et al. The probacterial effect of type I interferon signaling requires its own negative regulator USP18. Sci Immunol. 2018;3(27). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsoi LC, Spain SL, Ellinghaus E, Stuart PE, Capon F, Knight J, et al. Enhanced meta-analysis and replication studies identify five new psoriasis susceptibility loci. Nat Commun. 2015;6:7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Langley RG, Elewski BE, Lebwohl M, Reich K, Griffiths CE, Papp K, et al. Secukinumab in plaque psoriasis--results of two phase 3 trials. N Engl J Med. 2014;371(4):326–38. [DOI] [PubMed] [Google Scholar]
- 60.Chiricozzi A, Guttman-Yassky E, Suarez-Farinas M, Nograles KE, Tian S, Cardinale I, et al. Integrative responses to IL-17 and TNF-alpha in human keratinocytes account for key inflammatory pathogenic circuits in psoriasis. J Invest Dermatol. 2011;131(3):677–87. [DOI] [PubMed] [Google Scholar]
- 61.Albanesi C, Cavani A, and Girolomoni G. IL-17 is produced by nickel-specific T lymphocytes and regulates ICAM-1 expression and chemokine production in human keratinocytes: synergistic or antagonist effects with IFN-gamma and TNF-alpha. J Immunol. 1999;162(1):494–502. [PubMed] [Google Scholar]
- 62.Teunissen MB, Koomen CW, de Waal Malefyt R, Wierenga EA, and Bos JD. Interleukin-17 and interferon-gamma synergize in the enhancement of proinflammatory cytokine production by human keratinocytes. J Invest Dermatol. 1998;111(4):645–9. [DOI] [PubMed] [Google Scholar]
- 63.Noe MH, Shin DB, Doshi JA, Margolis DJ, and Gelfand JM. Prescribing Patterns Associated With Biologic Therapies for Psoriasis from a United States Medical Records Database. J Drugs Dermatol. 2019;18(8):745–50. [PMC free article] [PubMed] [Google Scholar]
- 64.Toh S, Li L, Harrold LR, Bayliss EA, Curtis JR, Liu L, et al. Comparative safety of infliximab and etanercept on the risk of serious infections: does the association vary by patient characteristics? Pharmacoepidemiol Drug Saf. 2012;21(5):524–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.