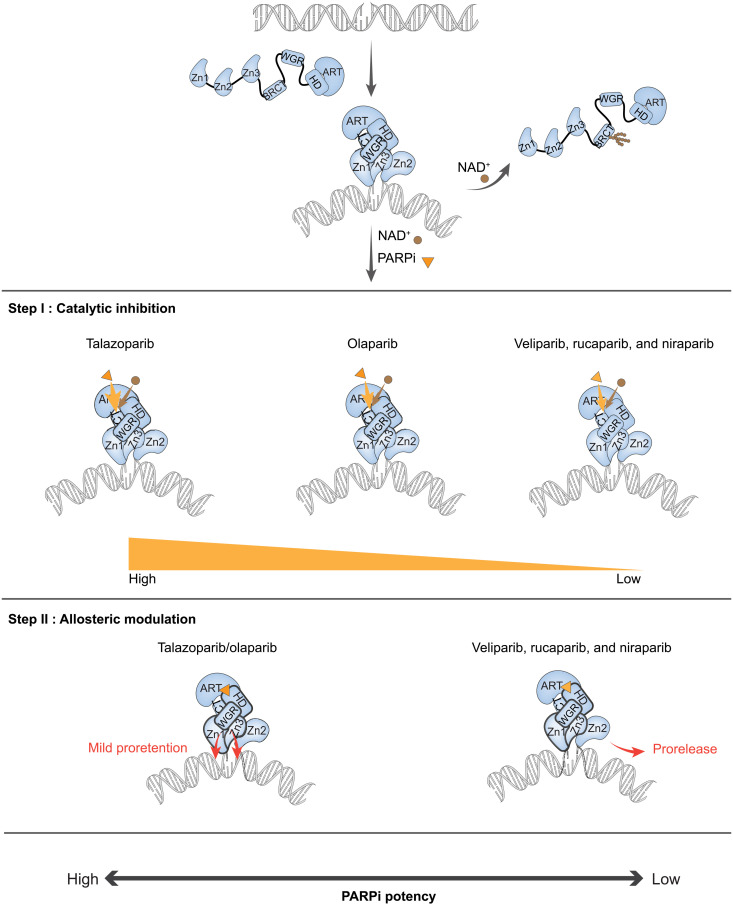
Fig. 5. A two-step model for clinical PARPi mediated PARP1 retention on DNA lesions.
NAD+ is depicted as a brown dot, and PARPi is depicted as an orange triangle. (i) PARP1 recognizes and binds to the DNA lesion through its N-terminal DNA binding domains, which activates its C-terminal domain polymerase activity to add ADP-ribose to its automodification domain. Extensive PARylation of PARP1 leads to release from DNA due to the high negative charge of poly-ADP ribose. (ii) Step I catalytic inhibition: PARPi retain PARP1 at the lesion by competing with NAD+ for the catalytic pocket binding site. (iii) Step II allosteric modulation: Once PARPi are bound at the catalytic pocket of PARP1, they further affect PARP1 retention on DNA by inducing allosteric modulation. PARPi mediate allostery through the Zn1-WGR-HD and WGR-Zn3-HD interfaces. Talazoparib and olaparib show mild proretention activity, while veliparib, rucaparib, and niraparib support prorelease.