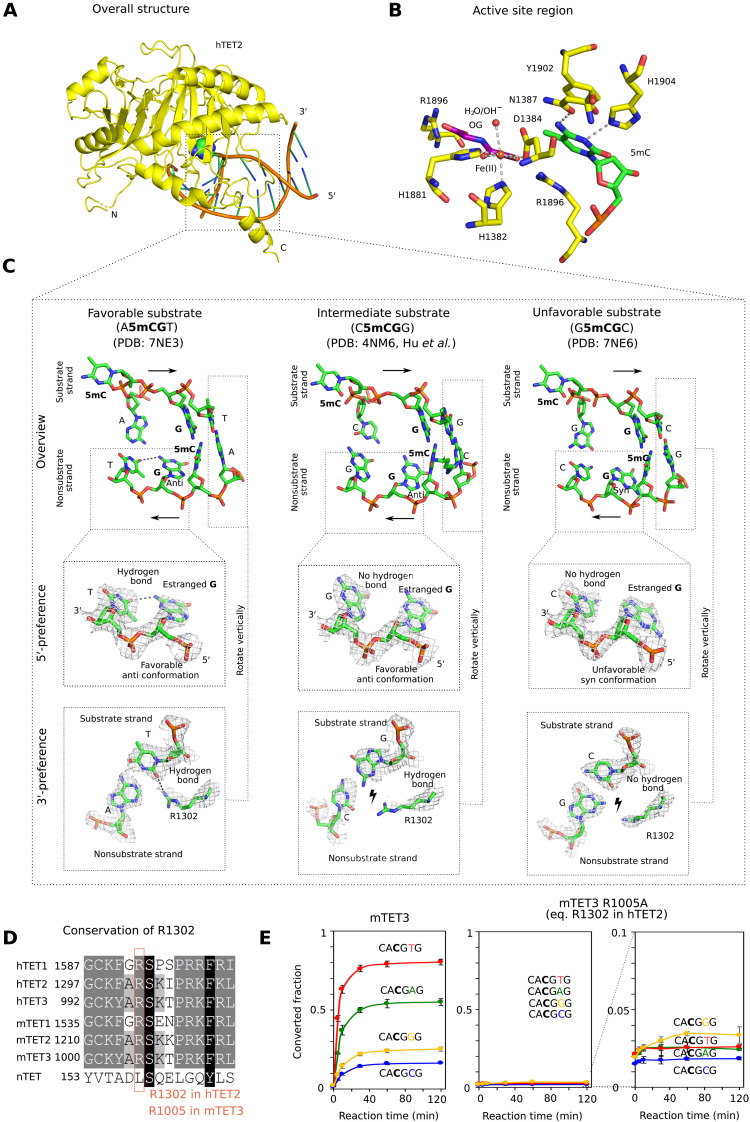
Fig. 3. Structural basis for TET sequence specificity.
(A) Structure of the core region of hTET2 (residues 1129 to 1936, with a 15-residue GS linker replacing disordered residues 1481 to 1843) with the most favorable substrate. Protein is shown in yellow in ribbon representation and DNA in schematic representation (brown backbone and green/blue nucleobases). The substrate 5mC base is highlighted in all-atom representation. The structure with the least favorable substrate is indistinguishable at this level of detail, except at the very N terminus, which is very uncertain because of high B factors (fig. S3). (B) Active site region with key hTET2 residues (yellow), Fe2+ (brown), the cosubstrate analog N-oxalylglycine (purple), and the substrate 5-methyl-2′-deoxycytidine monophosphate (green). At the level of resolution of the crystal structures, the active site regions are indistinguishable for the complexes with the most and least favorable substrates. (C) Conformation of the central four 2′-deoxynucleotides of substrate and nonsubstrate strands. In the magnified regions, composite omit densities contoured at 1σ are shown. (D) Conservation of the arginine residue (R1302 in hTET2) responsible for 3′-substrate preferences in the TET paralogs. (E) Confirmation of the relevance of the conserved arginine (R1302 in hTET2 and R1005 in mTET3) for the 3′-substrate preference. A synthetic substrate containing four 5mCG sites embedded in CA5mCGNG context differing only in the base pair immediately downstream of the methylated CG was subjected to oxidation by mTET3 or mTET3 R1005A, followed by quantification of conversion of 5mC to 5fC and 5caC by bisulfite sequencing.