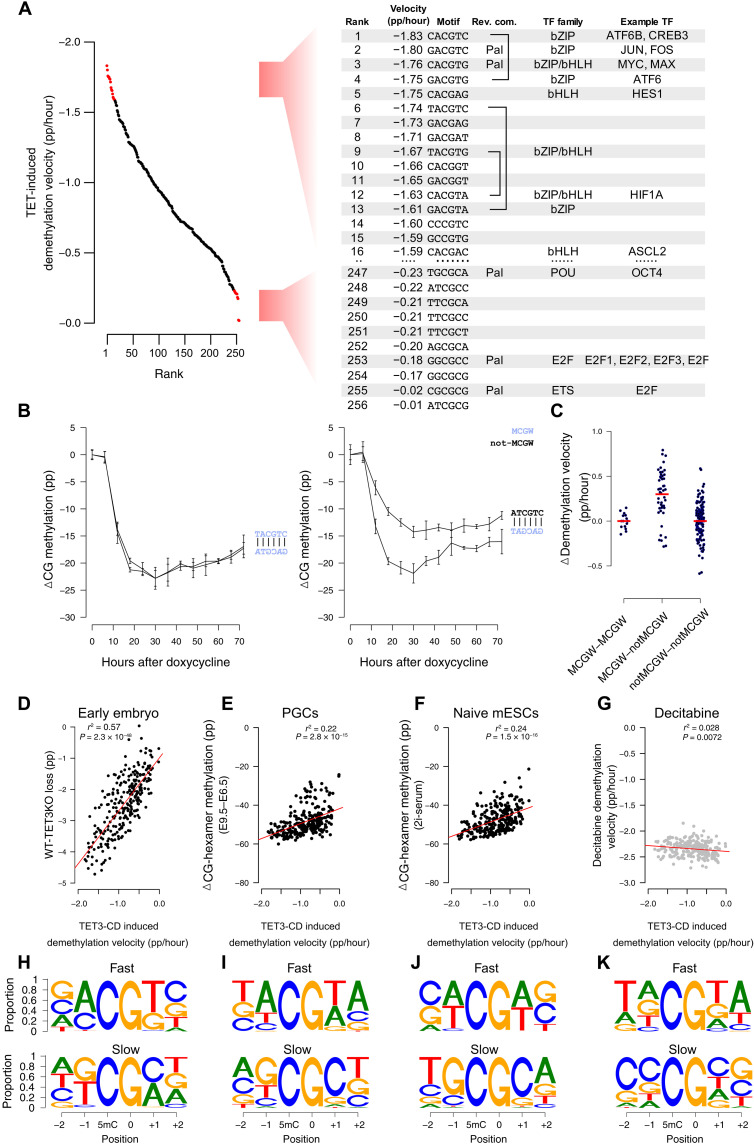
Fig. 6. TET selectivity in cultured cells coincides with methylation-sensitive transcription factors, is strand dependent, and correlates with global demethylation in vivo.
(A) Many fast demethylating CG-containing hexamer motifs following mTET3 expression (ranks 1 to 16) bind bZIP and bHLH methylation-sensitive transcription factors. Slow demethylating sites (ranks 247 to 256) bind methylation-sensitive E2F and POU family transcription factors. (B) Many complementary motifs have similar demethylation kinetics (e.g., TACGTC; left); however, some are significantly discordant (e.g., ATCGTC; right). (C) CG-containing hexamers with MCGW in one strand demethylate faster than complementary ones without MCGW (MCGW-notMCGW). In contrast, motifs where MCGW is present on both strands (MCGW-MCGW) or not (notMCGW-notMCGW) show equal demethylation rates. (D to G) CG-containing hexamer demethylation velocities following mTET3 overexpression (x axis, mTET3 induced demethylation velocity) and the demethylation found in mouse (y axis) (D) postfertilization embryos following TET3-knockout (WT-TET3KO), (E) primordial germ cells (E9.5 to E6.5), and (F) naive mESCs. Only a weak negative correlation exists with (G) decitabine-treated cells. (H to K) IMPP for the demethylating scenarios listed in (D) to (G). Shown is the proportion of nucleotides that are demethylated the fastest (top) and slowest (bottom) at each motif position when all other nucleotides in that motif are kept constant.