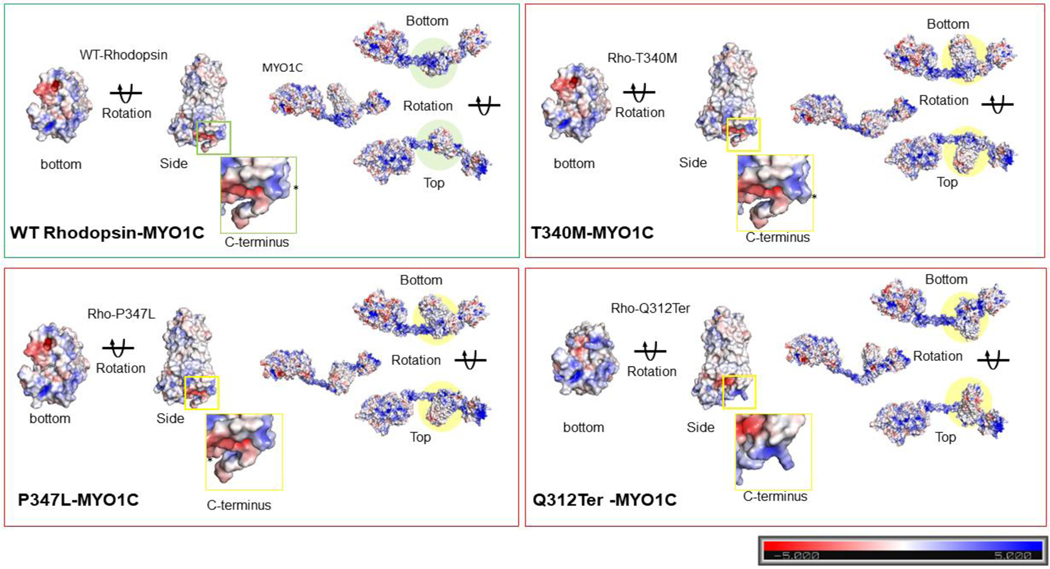
Figure 6: Electrostatic surface potential of individual WT and mutant Rhodopsin molecules and in complex with MYO1C.
The bar denotes and marks the surface colors fixed at −5 red and +5 blue, displaying the charge distribution in the surface map of the structure. The composition of amino acid residues determines the final charge distribution, hence it makes a significant contribution in the protein interactions. (A) The C-terminus overhang of WT-Rhodopsin has a conserved VXPX ciliary targeting motif, the panel shows the charge distribution of the entire amino acid residues accessible for cytosolic proteins interaction, on the bottom surface of Rhodopsin. The rotated 3D structure of WT-Rhodopsin shows a side view of it and the overhang with an asterisk symbol for the reference site. The MYO1C-WT Rhodopsin docking structure shows a charge distribution on the surface and the rotated image shows the alignment of Rhodopsin interaction under wild-type conditions. (B) The C-terminus overhang of Rho-T340M mutant has a conserved VXPX ciliary targeting motif in close proximity. The panel shows the charge distribution of the amino acid residues accessible for cytosolic proteins interactions on the bottom surface of T340M. The rotated 3D structure of T340M shows a side view of it and the overhang with an asterisk symbol for the mutation and sidechain of Leucine. The MYO1C-Rho T340M mutant docking structure shows a charge distribution on the surface with the rotated image are showing the alignment difference of T340M interaction. (C) The C-terminus overhang of Rho-P347L has a point mutation at the conserved VXPX ciliary targeting motif on the P residue. The panel shows the charge distribution of the amino acid residues accessible for cytosolic proteins interactions on the bottom surface of Rho-P347L. The rotated 3D structure of Rho-P347L shows a side view of it and the overhang with an asterisk symbol for the mutation and sidechain of Leucine. The MYO1C-P347L docking structure shows a charge distribution on the surface with the rotated image are showing the alignment difference of P347L interaction. (D) The C-terminus overhang deleted from Q312Ter mutation the conserved VXPX chain is not available. The panel shows the charge distribution of the amino acid residues accessible for cytosolic proteins interactions on the bottom surface of Q312Ter. The rotated 3D structure of Q312Ter shows a side view of it and with the reduced overhang. The MYO1C- Q312Ter docking structure shows a charge distribution on the surface with the rotated image are showing the alignment difference of Q312Ter interaction under mutant condition. The mutantRhodopsin structures generated from the mutagenesis wizard in PyMol.