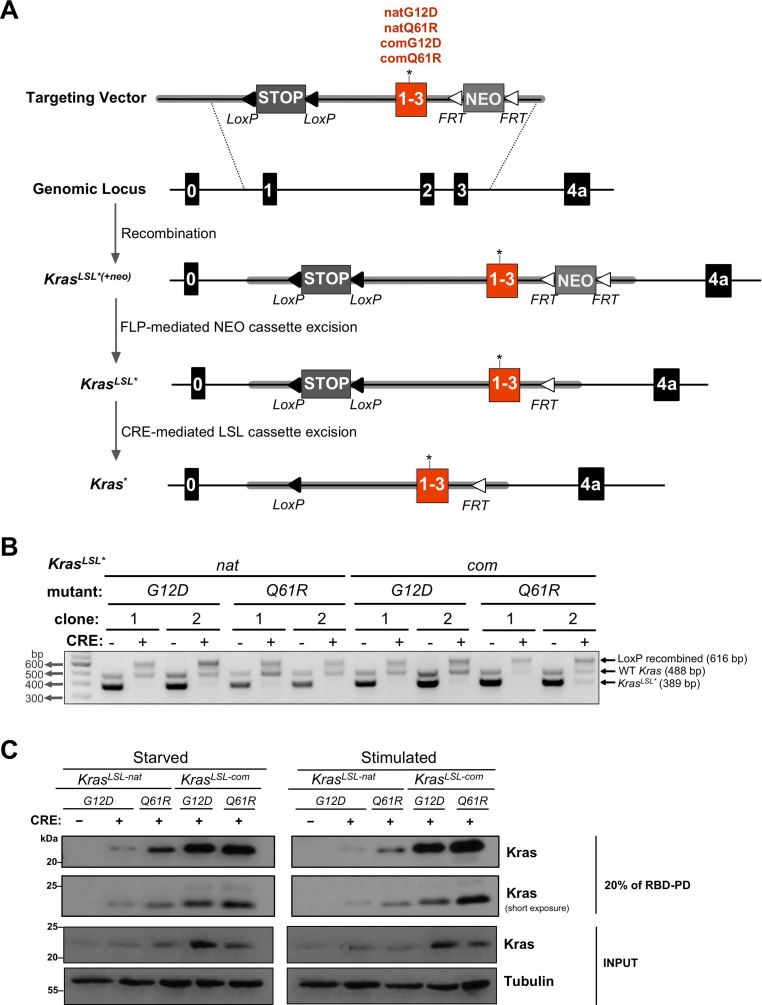
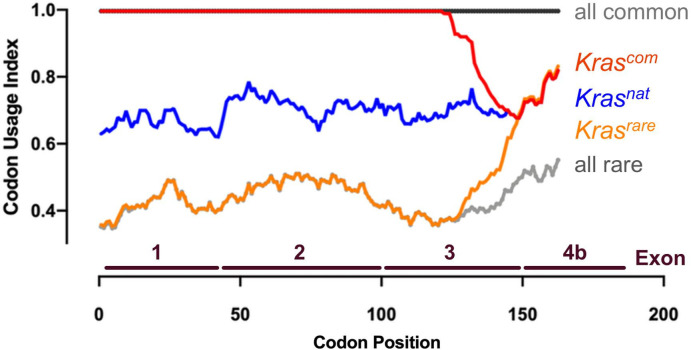
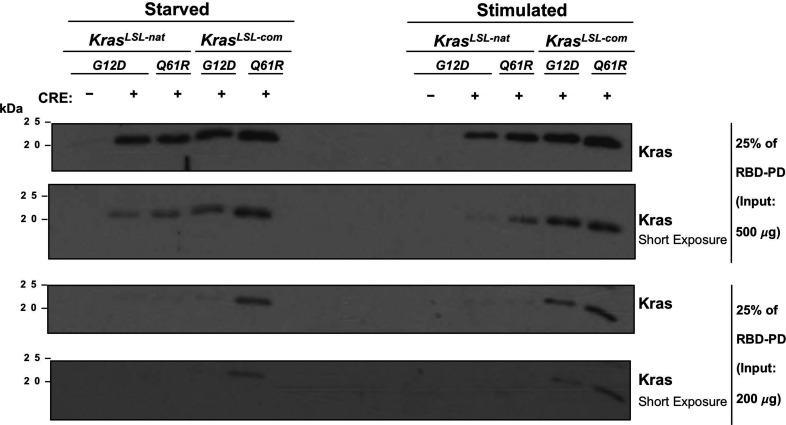
Figure 1. Conditional KrasLSL alleles with different oncogenic mutations and codon usage.
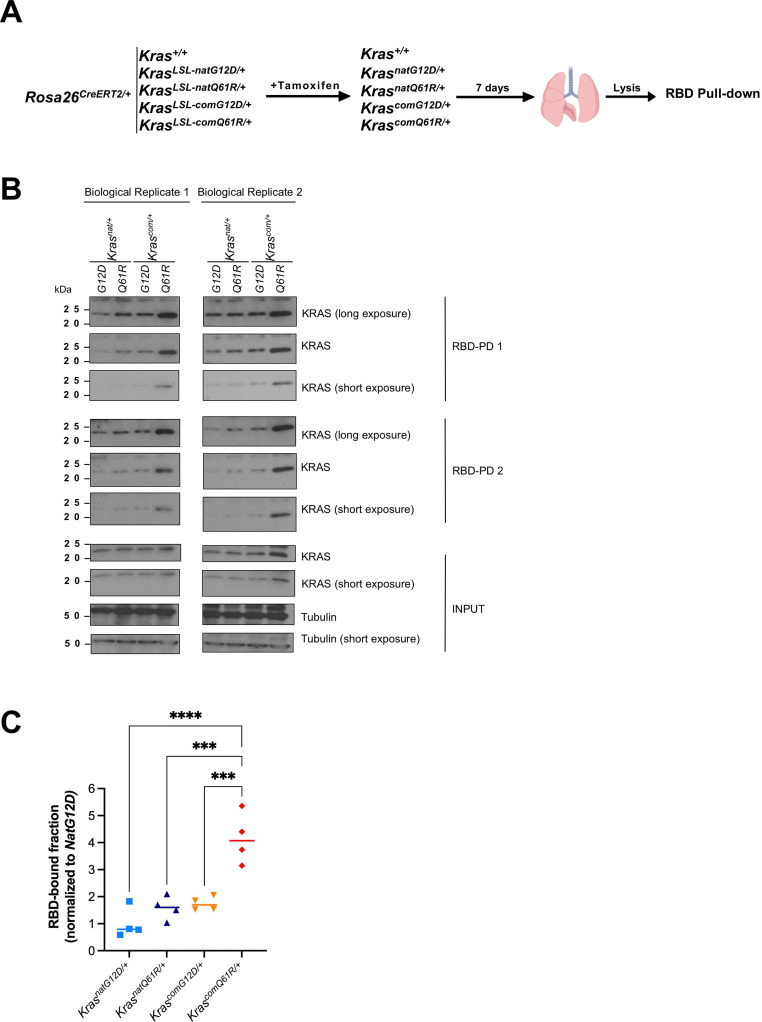
(A) Schematic of generating and activating KrasLSL alleles with the first three coding exons fused and encoded by native (nat) versus common (com) codons with either a G12D or Q61R mutation. (B) PCR genotyping of two independently derived mouse embryonic fibroblast (MEF) cultures (two biological replicates) with the indicated Kras alleles in the absence and presence of Cre recombinase (CRE) to detect the unaltered wild-type Kras allele product (WT, 488 bp) and the unrecombined (KrasLSL*, 389 bp) and recombined (LoxP recombined, 616 bp) KrasLSL allelic products. Gel images were cropped and color inverted for optimal visualization. Full-length gel images are provided in Figure 1—source data 2. (C) Expression levels, determined by immunoblot with an anti-Kras antibody, RAS activity levels, determined by RBD pull-down (RBD-PD) of lysates collected from MEF cells derived from mice with the indicated Kras alleles in the presence of Cre recombinase (CRE). MEF cultures derived from KrasLSL-natG12D/+ mice in the absence of Cre recombinase were used as negative control. MEF cultures were either serum starved overnight (starved) or serum starved overnight followed by serum stimulation for 5 min (stimulated). 20% of the elute from RBD-PD and 30 μg total protein from the total cell lysates were loaded. Tubulin serves as loading control. One of two biological replicates; see Figure 1—figure supplement 3 for the second biological replicate. Full-length gel images are provided in Figure 1—source data 3.