Figure 2.
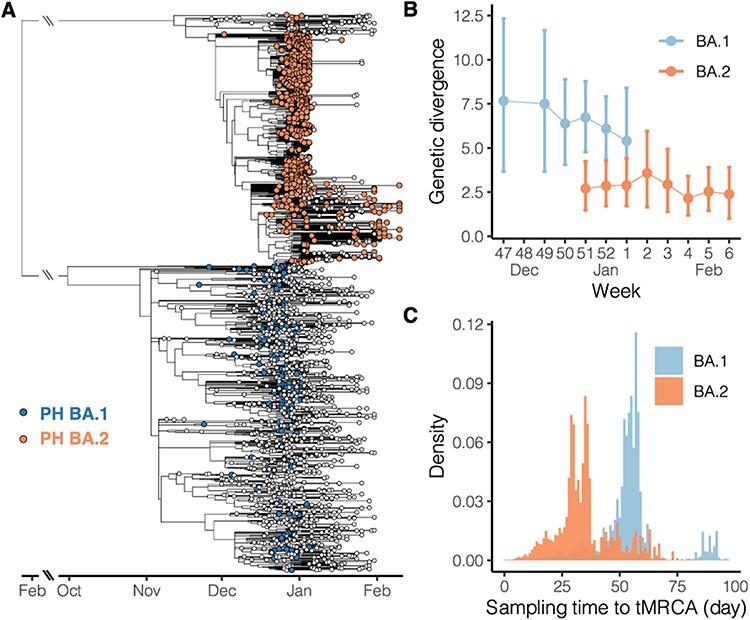
Phylogenetic relationship of SARS-CoV-2 Omicron variants isolated in the Philippines. (A) Time-scaled tree was inferred by TreeTime using Omicron variant genome sequences isolated in the Philippines and from the global database. The blue and orange tips indicate the BA.1 and BA.2 lineage viruses, respectively, isolated in the Philippines, whereas the white tips indicate the viruses isolated in the other countries. Long branches descending from the common ancestor of BA.1 and BA.2 are shortened. (B) Average genetic divergence of viruses isolated in the Philippines. Error bars represent 95 per cent bootstrap percentiles. (C) Distribution of time intervals from tMRCA to the isolation time. The time intervals were calculated based on pairs of Philippine taxa on the time-scaled tree (A); for each pair, the larger time difference was recorded.
