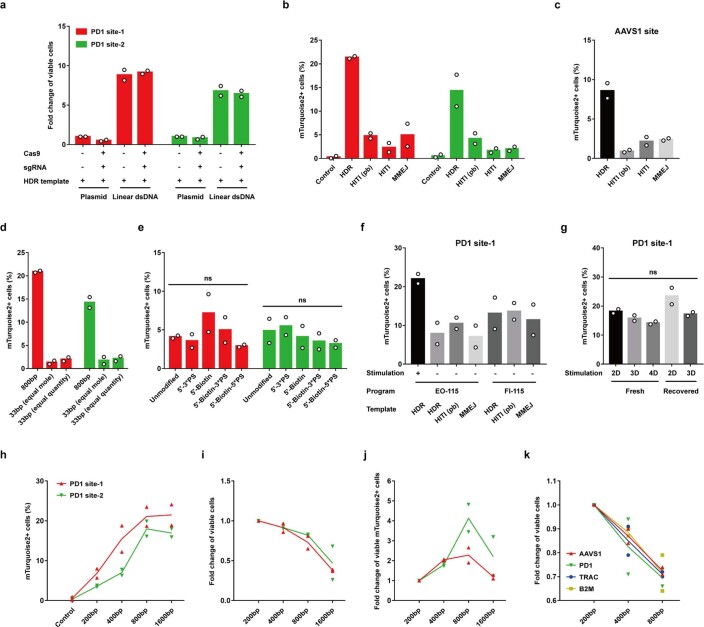
Extended Data Fig. 1. Optimization of the conditions for producing non-viral gene-specific targeted T cells.
a-j, The sequence of the fluorescent protein mTurquoise2 was used as a target to optimize the conditions for generating non-viral gene-specific integrated T cells. a, Number of viable cells calculated 7 days after electroporation by using different protocols. Equal quantities of circular plasmid DNA and linear double-stranded DNA (dsDNA) were used. Due to acquisition of higher cell viability, templates in the form of linear dsDNA were chosen for all the following experiments. b-c, Homologous recombination efficiency of mTurquoise2 at two PD1 sites (b) and one AAVS1 site (c) by using different DNA templates. HDR, homology directed repair. HITI, homology-independent targeted integration. HITI (pb), HITI template with 50bp protection base pairs flanking the target sequence. MMEJ, microhomology-mediated end joining. d, Homologous recombination efficiency of mTurquoise2 using 33bp or 800bp homology arms. Equal mole or quantity of template harboring 33bp homology arms was used, compared with template with 800bp homology arms. e, Homologous recombination efficiency of mTurquoise2 by using unmodified or modified DNA templates with 200bp homology arms. PS, phosphorothioate. Biotin was modified at the first base pair from the 5’ side. PS was modified at the first three or five base pairs from the 5’ side. f, Homologous recombination efficiency of mTurquoise2 in unstimulated or stimulated T cells using different electroporation programs and DNA templates. g, Homologous recombination efficiency of mTurquoise2 in fresh or recovered T cells after stimulation for indicated days by using HDR templates with 800bp homology arms. h-j, Homologous recombination efficiency (h) and numbers of all viable cells (i) and viable mTurquoise2+ cells (j) were detected using mTurquoise2 templates with different homology arm lengths. k, Number of all viable cells was enumerated using CAR templates with different homology arm lengths. 800bp and 20bp homology arms were used in HDR and MMEJ templates, respectively (a–c, f). Equal moles of DNA template were used in b, c, e–k. The homologous recombination efficiency was determined 7 days after electroporation in b-k. All the experiments were performed in cells from two independent healthy donors. Mean value is shown in all the figures. P values were calculated by one-way ANOVA with Tukey’s multiple comparisons test (g) or two-way ANOVA with Tukey’s multiple comparisons test (e).