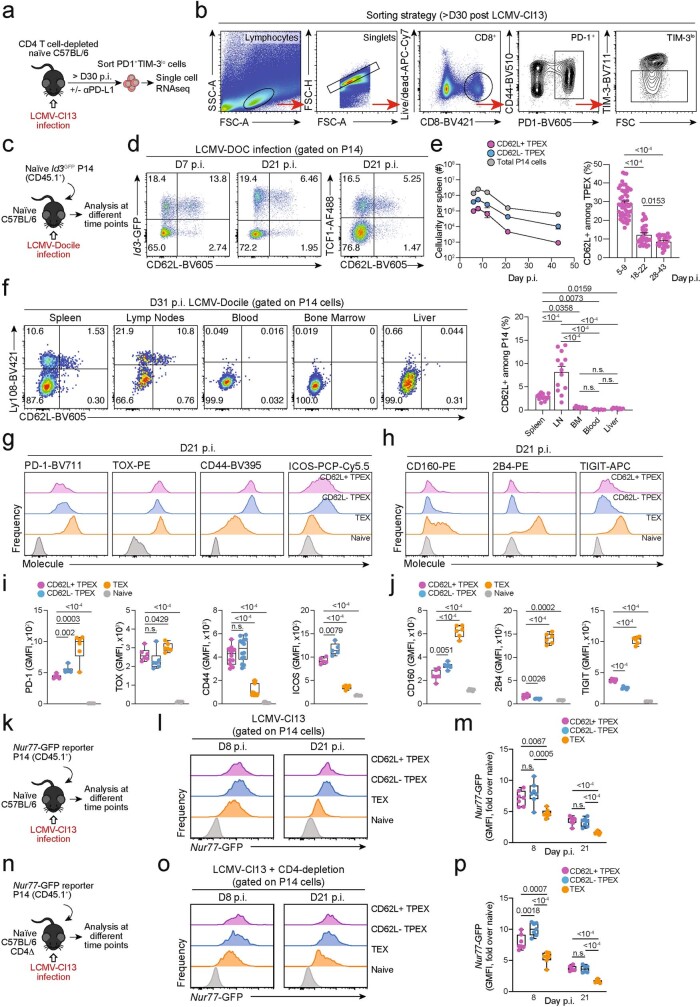
Extended Data Fig. 1. Isolation of polyclonal exhausted T cells for scRNA-seq and phenotypic characterization of CD62L+ TPEX cells in chronic infection.
(a, b) CD4+ T cell-depleted naive mice were infected with LCMV-Cl13, treated with or without anti-PD-L1, and exhausted PD-1+TIM-3lo T cells were sorted at >day 30 post-infection as described19. (a) Schematic of the experimental set-up. (b) Flow cytometry plots showing the sorting strategy. (c–j) Naive congenically marked (CD45.1+) Id3-GFP P14 cells were transferred to naive recipients (Ly.5.2), which were then infected with LCMV-Docile. Splenic P14 T cells were analysed at the indicated time points after infection. (c) Schematic of the experimental set-up. (d) Flow cytometry plots showing the expression of Id3-GFP, TCF1 and CD62L among splenic P14 T cells at 7 and 21 dpi. (e) Quantification showing absolute numbers of splenic CD62L+ TPEX, CD62L− TPEX and total P14 cells (left) and frequencies of CD62L+ cells among TPEX cells (right) at the indicated time points after infection (f) Flow cytometry plots showing the expression of Ly108 and CD62L and quantification of CD62L+ TPEX cells among P14 T cells in the spleen, lymph nodes, blood, bone marrow and liver at day 31 post LCMV-Docile infection. (g–j) Histograms (g, h) and quantification (i, j) of expression of molecules as indicated in P14 T cell subsets and naive CD8+ T cells. (k–p) Congenically marked naive Nur77-GFP reporter P14 T cells were transferred into naive (k–m) or CD4-T-cell-depleted (n–p) recipient mice, which were subsequently infected with LCMV-Cl13. Nur77-GFP expression was analysed at indicated time points post-infection. (k, n) Schematics of the experimental set-up. Histograms (l, o) and quantifications (m, p) showing Nur77-GFP expression in the indicated P14 T cell subsets at 8 and 21 dpi. GMFI, geometric mean fluorescence intensity. Dots in graphs represent individual mice; box plots indicate range, interquartile and median; horizontal lines and error bars of bar graphs indicate mean and s.e.m. Data are representative of two independent experiments (e, f, i, j) and all analysed mice (m, p). P values are from two-tailed unpaired t-tests (e, i, j), two-way ANOVA (f), and one-way ANOVA (m, p); P > 0.05, not significant (n.s.).