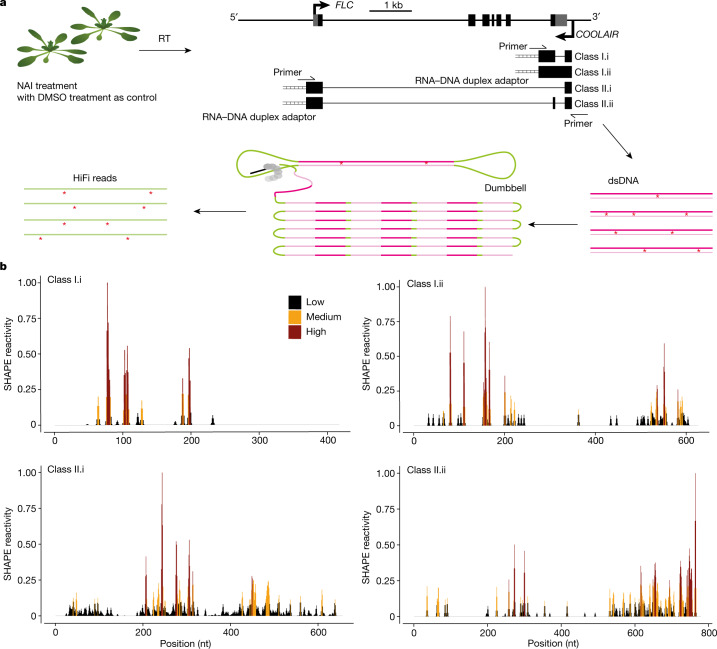
Fig. 1. smStructure-seq captures RNA secondary structure information of different transcript isoforms.
a, Schematic of the smStructure-seq design for RNA secondary structure probing of each COOLAIR isoform. The Arabidopsis seedlings were treated with NAI ((+)SHAPE) or DMSO ((−)SHAPE). Total RNA was extracted, and the RNA–DNA hybrid adaptors (ladder symbol) were added to the reverse-transcription (RT) reaction using TGIRT-III enzyme. dsDNAs were generated by adding specific primers for all of the COOLAIR isoforms. The dumbbell adaptors were then ligated to the resulting dsDNAs to generate PacBio libraries. The raw subreads were converted to high-accuracy HiFi reads (or circular consensus sequences)14 to generate the mutation rate profiles. b, The normalized SHAPE reactivities derived from the mutation rate profiles were plotted for different class I (under cold-grown conditions) and II (under warm-grown conditions) COOLAIR transcript isoforms. The normalized SHAPE reactivity is calculated from merged n = 2 biological replicates. These reactivity values are colour-coded and shown on the y axis.