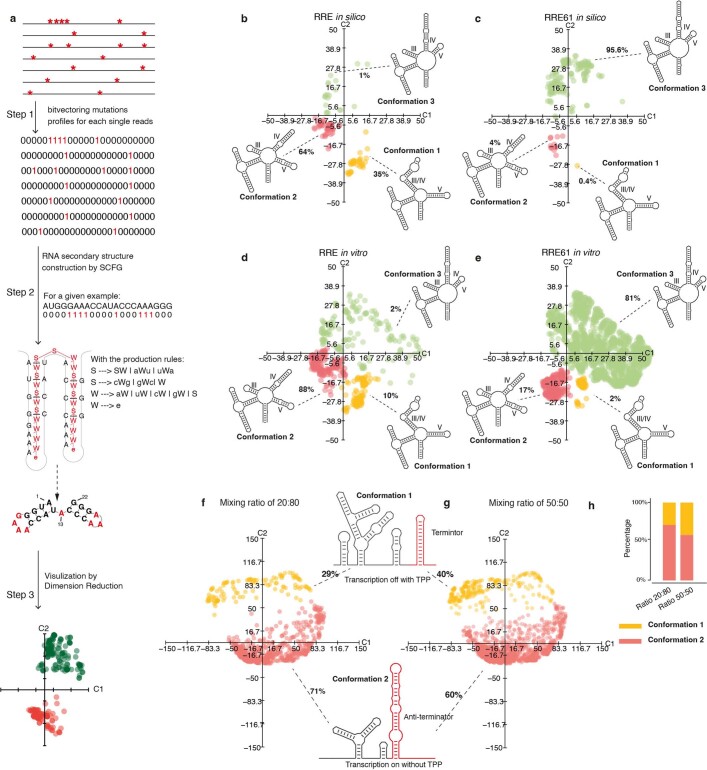
Extended Data Fig. 2. DaVinci conformation analysis pipeline and its validation.
a, DaVinci conformation analysis pipeline. Each line is referring to one sequencing read. The red stars denote the mutations including mismatch and deletions. In step 1, the sequencing reads are bit-vectorized following the rules: “0” if a base is wild type and “1” if the base is mutated. In step 2, SCFG42 was applied to derive the RNA structures that can best-represent each mutation profile. For example, given sequence “AUGGGAACCAUACCCAAAGGG” with a bitvector of “00011100001000111000”, the production rule (showed in the step 2) can derive the RNA structures as showed in step 2, independent of thermodynamic parameters. “|” in the rules represents the logic of “or” between production rules. The red “1”s or letters indicate the mutation information or single-stranded nucleotides. In step 3, the collected RNA structures derived from each individual mutation profile were transformed into numeric matrix of RNA structure element and subjected to dimensionality reduction. Then, the representative RNA structures for each conformational cluster were determined. Detailed description was in the Methods section. b–e, The in silico (b, c) and in vitro (d, e) RNA conformational landscape of HIV-1 Rev response element (RRE) region in wild-type sequence (RRE) or mutant RRE61. f, Davinci-determined RNA conformation landscape for TenA RNAs folded with and without TPP ligands. The folded RNAs were probed in vitro and pooled with the ratio of 20 (TPP-treated RNAs):80 (non TPP-treated RNAs). g, Similar to (f) but with the pooling ratio of 50 (TPP-treated RNAs):50 (non TPP-treated RNAs). The detailed discussions of (b–g) were in Supplementary Discussion. h, Proportions for each cluster detected by DaVinci. The ratios are derived from (f, g).