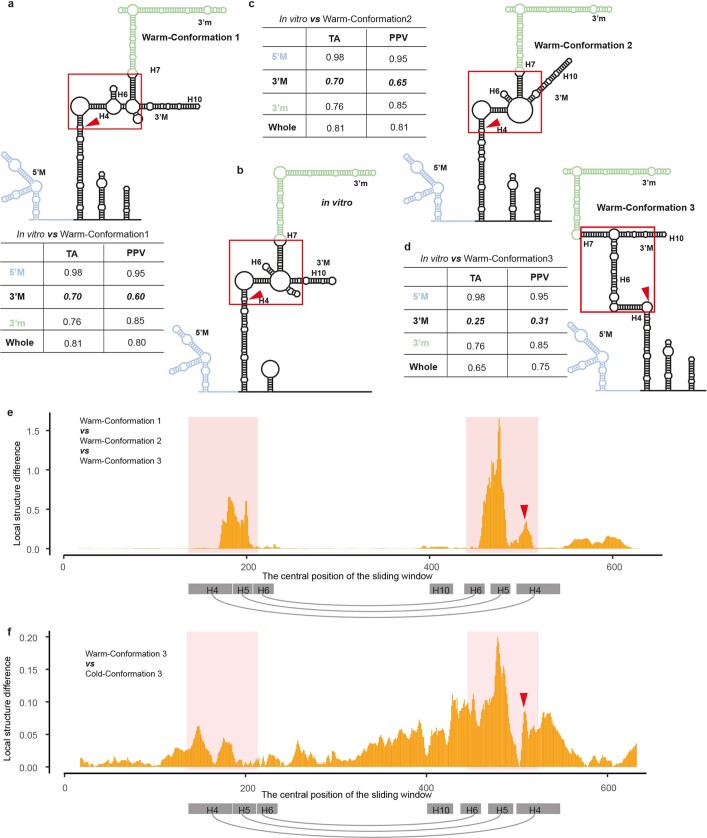
Extended Data Fig. 4. Identification of hyper-variable RNA structural region.
a, RNA structure model of warm conformation 1 in (Fig. 2a). The red triangle indicates the site corresponding to FLC TSS. The table is the similarity comparison between each domain as well as the whole structure of warm conformation 1 and the previously reported in vitro5 RNA structure (b). The topological similarity was based on the tree alignment (TA)43 and calculated by RNAforester35. The base-pairing similarity was calculated using positive predictive value (PPV)44. The red square is showing the dramatic structure change between in vitro and in vivo RNA structure. b, RNA structure model of previously reported in vitro5 Class II.i RNA structure. c, d, RNA structure models of warm conformations 2 and 3 in (Fig. 2a). The similarity comparison of TA and PPV were listed in the table respectively. The central 3’ M domain showed the lowest topological and base-pairing similarities (bold and oblique) between in vitro Class II.i and warm conformations 1, 2 and 3. e, The local structural difference was measured by the -log10(P value) in two-way ANOVA test of single-strandedness among these three warm conformations in a sliding window of 30 nt. The red shadow region indicated the greatest difference, i.e, hyper-variable region, among warm conformations 1, 2 and 3. The red shadow regions are corresponding to the red squares in (a–d). The sequences in H4, H5 and H6 helix were draw in grey rectangle with grey arch indicating the helix formation. f, The local structural difference was measured by the -log10(P value) in the t-test of single-strandedness between the warm specific conformation (warm conformation 3) and cold specific conformation (Cold-Conformation 3). The red shadow regions are corresponding to the red squares in (a–d).