Figure 1.
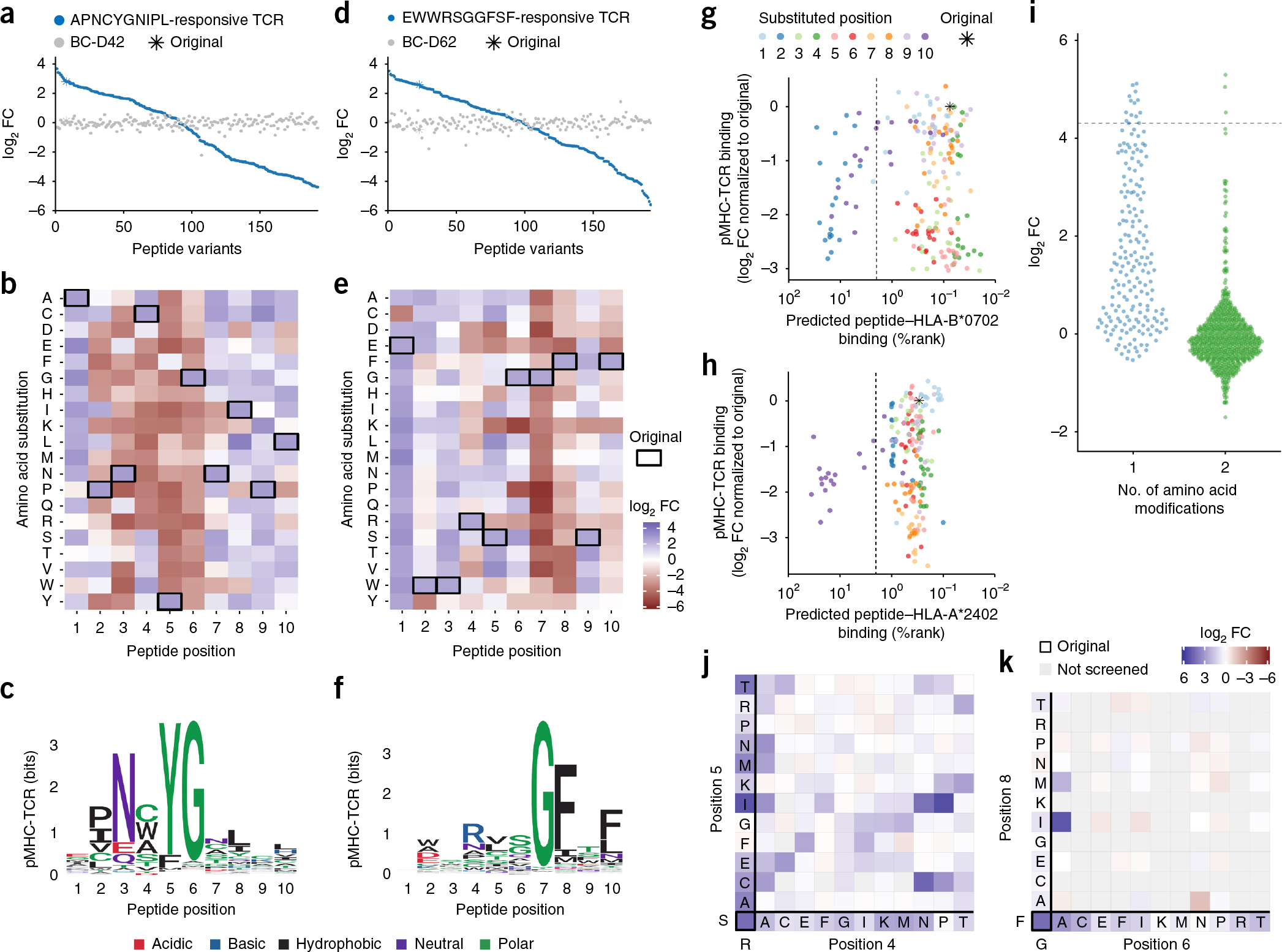
The fingerprints of two different TCRs that recognize MCC-derived peptides restricted to HLA-B*0702 or HLA-A*2402. (a–c) Results obtained from the DNA barcode-based analysis of T cells transduced with a TCR recognizing the HLA-B*0702 -restricted peptide APNCYGNIPL. The analysis was performed with all possible variations of peptides created by single-position amino acid substitutions. (a) The hierarchy of pMHC interactions expressed as log2FC of read counts relative to a triplicate baseline sample (see Supplementary Note 1). A healthy donor PBMC sample (BC-D42) was screened with the same MHC multimer panel in parallel. For both samples, the plotted order of log2FCs of each pMHC-associated DNA barcode is determined by the hierarchy obtained from screening the HLA-B*0702APN-responsive TCR. (b) Heat map of amino acid preferences of the HLA-B*0702APN-responsive TCR based on data from a. Each row represents a given amino acid and each column a position in the peptide sequence. The amino acids of the original peptide target are marked in black boxes. (c) Recognition pattern of the HLA-B*0702APN-interacting TCR, here visualized as a sequence logo based on the data from a and b. (d–f) Results obtained from the DNA barcode-based analysis of T cells transduced with a TCR recognizing the HLA-A*2402-restricted peptide EWWRSGGFSF. The analysis was performed with all possible variations of peptides created by single-position amino acid substitutions. Visualization of data corresponds to a–c. b and e are colored according to the same key. (g,h) Scatter plot of the predicted peptide binding, percentage rank (%rank, x axis) of all naturally occurring amino acid substitutions of APNCYGNIPL to HLA-B*0702 (g) or EWWRSGGFSF to HLA-A*2402 (h), in relation to the experimentally obtained TCR–pMHC interaction (y axis). The color indicates the position of the amino acid substitution. %rank < 2 (dotted line) marks the recommended cutoff of peptides that are considered binders to MHC. (i–k) Results from a parallel MHC multimer analysis of the TCR recognizing the HLA-A*2402 restricted peptide EWWRSGGFSF with a MHC multimer library composed of peptides with single amino acid substitutions corresponding to the one used in d–f, as well as double amino acid substitutions covering 12 naturally occurring amino acids, where positions 4–8 are substituted two amino acids at a time (n = 967; see full list in Supplementary Data 4 and 7). (i) The obtained log2FC values, grouped according to the number of substitutions, one (n = 191) or two (n = 776), within the peptide sequence. Dotted line at 4.30 indicates the original peptide. (j,k) Heat maps showing the log2FC obtained for peptides with amino acids substituted at (j) positions 4 and 5 simultaneously or (k) positions 6 and 8 simultaneously. Each row and column represents a given amino acid substitution (see heat map of all screened substitutions in Supplementary Fig. 9). The original amino acids are marked in bold and peptides in the same row or column are substituted at only one position, indicated with the one-letter code. j and k are colored according to the same key. All data are representative of duplicate analyses.
