Fig. 2: eCLIP demonstrates sequence-specific binding of GATA4 to RNA in human iPS-CPs.
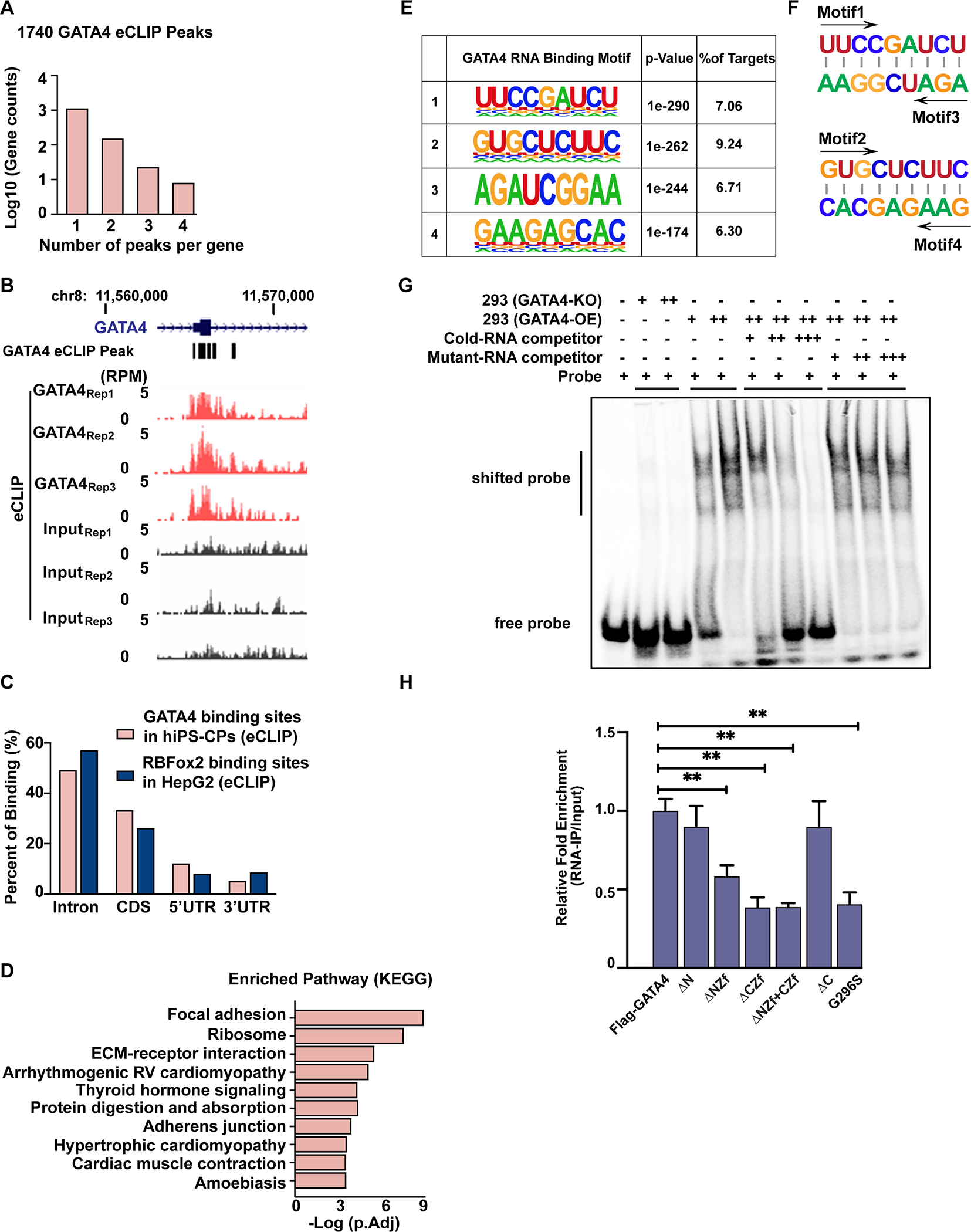
(A) Numbers of GATA4-eCLIP peaks in human iPS-CPs. (B) Integrated genome viewer tracks centered on the GATA4 gene. OmniCLIP identified peak regions indicated as black boxes above. (C) Distribution of GATA4-eCLIP (hCPCs) binding sites or RbFOX2-eCLIP (HepG2) binding sites across different regions of gene bodies. (D) KEGG pathway analysis of GATA4-bound RNAs. (E) Weblogos depicting the most significantly enriched GATA4-eCLIP-binding motifs by eCLIP in human iPS-CPs. (F) The motifs 1 or 2 were the reverse complementary sequences of motifs 3 or 4, respectively. (G) EMSAs were performed by adding GATA4-knockout (KO) or GATA4-overexpressing (OE) HEK293T cell lysate to an IRDye 800 labeled RNA probe containing the GATA4-binding motif 4 sequence. EMSA was performed by incubating HEK293T GATA4-OE lysate with labeled probe in the presence of 200(+)-, 1000(++)- and 2000(+++)-fold molar excess of the unlabeled competitor RNA or unlabeled mutant-RNA competitor. (H) RNA-pull down assay of GATA4 with the GATA4 binding site on CESR2 mRNA for GATA4. Immunoprecipitated mRNA were reverse-transcribed and amplified by qRT-PCR. One-way ANOVA coupled with a Tukey test was used to assess significance. **P < 0.001.
