Fig. 4: GATA4 regulates mRNA splicing through direct interaction with mRNAs.
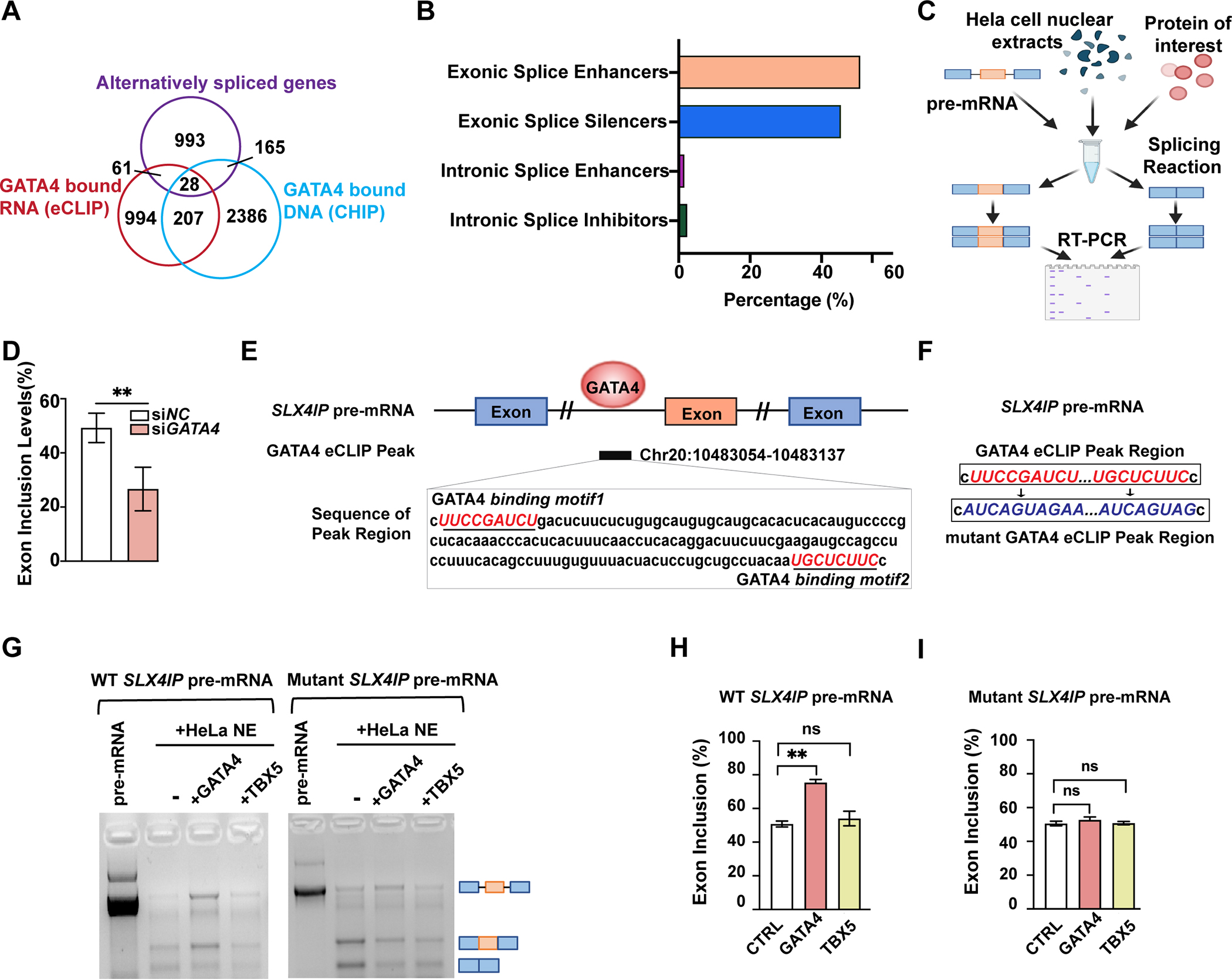
(A) Intersection of alternatively spliced genes upon GATA4-knockdown (KD) defined by rMATs (FDR <0.05, junction counts ≥5), genes with GATA4-eCLIP tag, and genes with GATA4-CHIP tag. (B) Quantification of GATA4 function in regulating splicing of 89 genes that were differentially spliced upon GATA4 KD and were bound to GATA4 based on an eCLIP binding peak. (C) Schematic representation of the in vitro splicing assay. (D) Exon inclusion level of significantly changed exon 3 of SLX4IP upon use of GATA4 siRNA compared to negative control (NC) siRNA. Data are shown as means ± SEM (n=4). Statistical significance was assessed using Student’s t test (**P < 0.001). (E) Integrated genome viewer tracks centered on gene SLX4IP. OmniCLIP identified peak regions indicated as black box. GATA4 binding motifs indicated in red. (F) Sequence information of the WT or mutant SLX4IP pre-mRNA template used in in vitro splicing assay. (G) In vitro splicing of SLX4IP minigene reporter transcripts in Hela nuclear-extracts (NE), with or without the addition of GATA4 or TBX5 protein. Bands indicate sizes of splicing events schematized with pre-mRNA length versus two alternatively spliced events. (H and I) Quantification of relative exon inclusion level in (G). Data are shown as means ± SEM (n=3). One-way ANOVA coupled with a Tukey test was used to assess significance. ns., non-significant; **P < 0.001.
