FIGURE 2.
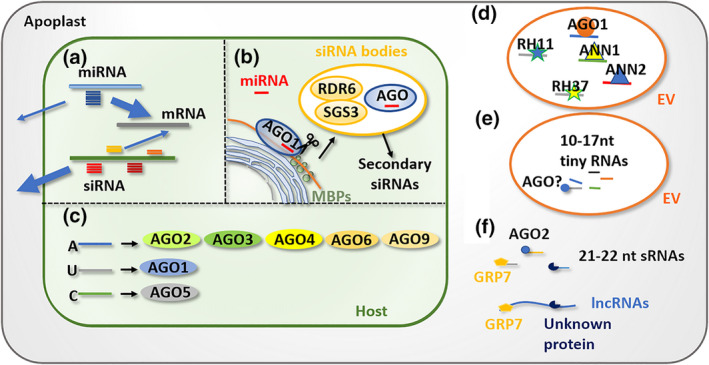
Potential mechanisms involved in small noncoding RNA (sRNA) sorting. (a) Plant microRNAs (miRNAs) with unique sequences are retained in the cytoplasm to regulate intracellular gene expression, while most small interfering RNAs (siRNAs), without intracellular targets, are more prone to be exported to the extracellular space. (b) miRNAs and siRNAs are synthesized in different cellular compartments. miRNA precursors are processed in the nucleus by Dicer‐like proteins. siRNA‐producing transcripts are cleaved on membrane‐bound polysomes (MBP) and rough endoplasmic reticulum, and further processed in siRNA bodies. The partitioning of sRNA biosynthesis potentially determines secretion of different classes of sRNAs. (c) sRNAs with the 5′ terminal nucleotides A, U, and C are preferentially recruited by AGO2/3/4/6/9, AGO1, and AGO5, respectively. sRNA loading by different AGOs may contribute to the selection of secreted sRNAs. (d) Extracellular RNAs encapsulated within extracellular versicles (EVs) need to be associated with RNA‐binding proteins for the purpose of sRNAs loading and/or stabilization. (e) Tiny RNAs (10–17 nucleotides [nt]) are enriched in EVs and could be selected by AGOs carried in EVs. (f) sRNAs (21–22 nt) and long noncoding RNAs (lncRNAs) are more probably translocated into the apoplast independent of EVs. Alternatively, AGO2, GRP7, and other RNA‐binding proteins are associated with RNAs to confer selection and/or stabilization.
