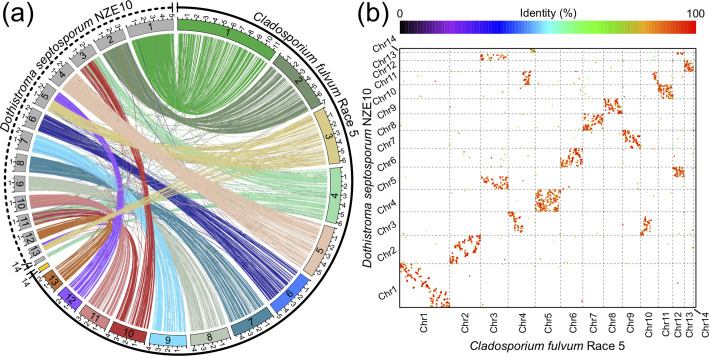
Fig. 6.
Mesosynteny between Cladosporium fulvum Race 5 and Dothistroma septosporum NZE10. The figure shows the whole-genome alignment produced with PROmer based on the six-frame translation of the genomes of C. fulvum Race 5 and of D. septosporum isolate NZE10. (a) Circos plot showing the collinearity between the chromosomes of the two species. Ribbons are based on nucleotide identity. (b) Dot-plot showing syntenic relations between the chromosomes of the two species. Chromosomes are numbered and dots are color-coded for percent nucleotide identity. The plots show a pattern of mesosynteny between C. fulvum Race 5 and D. septosporum NZE10, in which gene content is largely conserved within chromosomes, with few interchromosomal rearrangements, whereas gene order is not conserved. The alignment with the D. septosporum NZE10 genome produced high identity values, as shown in panel (b), which contrasts with the large difference in genome size compared to C. fulvum Race 5, as shown in the circos plot in panel (a). The mini-chromosome 14 (Chr14) of C. fulvum Race 5 had no matches with the genome of D. septosporum NZE10, supporting the hypothesis that this chromosome is dispensable. Chromosomes were named based on their size, from the longest to the smallest one, and are shown in scale in the circos plot, with tick labels indicating Mb. Whole-genome alignments of C. fulvum Race 5 with other Capnodiales are shown in Fig. S14.