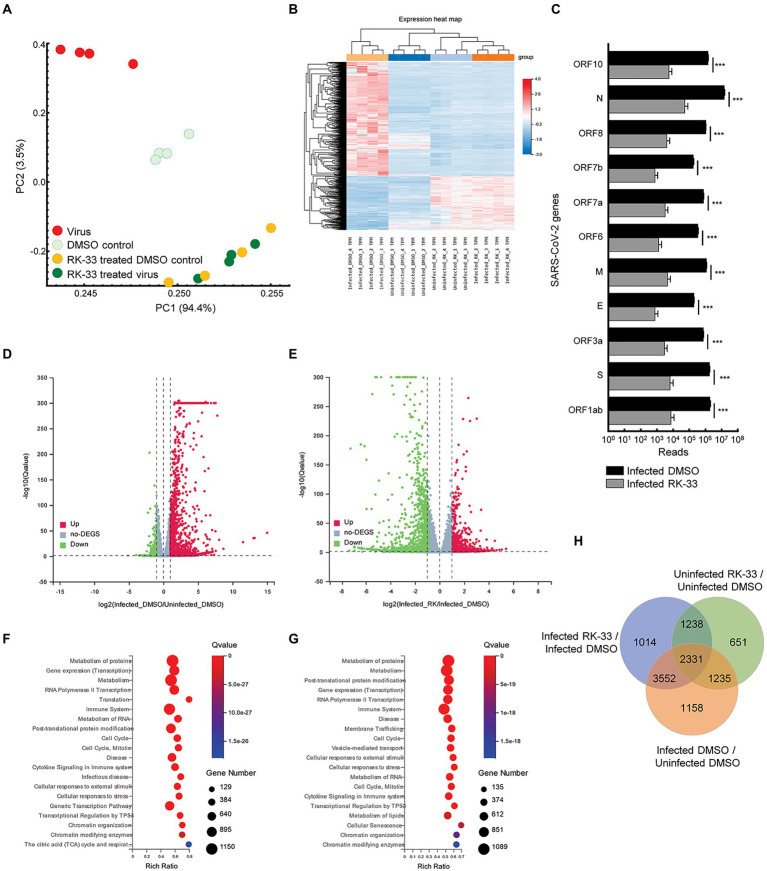
Figure 1.
Transcriptomics of viral infected Calu-3 cells. (A) PCA plot of RNA-seq data. PC1 accounts for 94% of the variation between the sample clusters. (B) Dendrogram and Heat map of RNA-seq data showing relationship between the various samples. Dendrogram was constructed using z-scores of TPM values of all RNA-seq sample reads. Horizontal sorting was by cluster order. (C) Histogram of SARS-CoV-2 RNA that are downregulated by RK-33 treatment of virally infected Calu-3 cells. *p < 0.05, **p < 0.005, ***p < 0.0005. (D) Volcano plot displaying effect of virus infection on Calu-3 cells. Genes differentially regulated in virally infected untreated samples were compared to uninfected samples. The X-axis represents the log2 fold change of the difference, and the Y-axis represents the-log10 significance value. Red represents DEG upregulated >2-fold change, green represents DEG downregulated, and gray represents non-DEGs. (E) Volcano plot displaying effect of RK-33 on virus infected Calu-3 cells. (F) Bubble plot displaying significantly enriched (Q < 0.05) Reactome pathways affected by SARS-CoV-2 infection of Calu-3 cells. X-axis displays the enrichment ratio which indicates more enrichment toward the right. Larger bubbles indicate more genes. (G) Reactome enrichment pathways affected by RK-33 treatment of virally infected Calu-3 cells displayed as a bubble plot. (H) Venn diagram displaying the number of genes dysregulated by SARS-CoV-2 infection, RK-33 treatment of uninfected cells, and RK-33 treatment of infected cells.