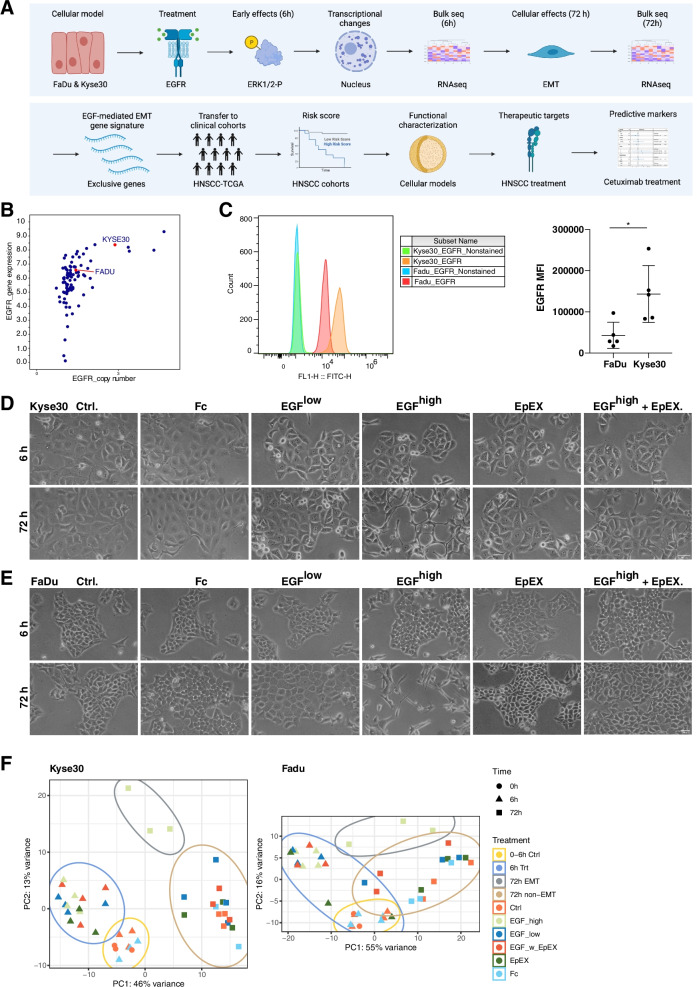
Fig. 1.
RNA-seq analysis of EGFR-mediated EMT. A Workflow: Kyse30 and FaDu cells were treated with EGF and EpEX inducing proliferation or EMT, respectively. Early transcriptional changes were assessed after 6 h and transcriptional differences associated with cellular effects after 72 h. EGFR-mediated EMT-associated genes serve to define prognostic gene signatures in clinical cohorts. Potential therapeutic and predictive markers in the EGFR-mediated EMT gene signature are explored via functional characterization and bioinformatic approaches. B Copy number variation and gene expression values of cell lines of the upper aerodigestive tract (esophageal carcinoma and HNSCC) were extracted from CCLE and are depicted as dot plot. C Kyse30 and FaDu cells were stained with EGFR-specific antibodies in combination with FITC-labeled secondary antibody. Shown are representative histograms (left panels) and mean expression values with SD of n = 5 independent experiments; * p-value < 0.05 (t-test). D-E Representative cytological pictures of Kyse30 and FaDu cells treated with the indicated components at 6 h and 72 h are shown (n = 4 independent experiments). Ctrl.: control treatment under serum-free conditions; Fc: recombinant immunoglobulin Fc region; EGFlow: 1.8 nM EGF; EGFhigh: 9 nM EGF; EGFhigh + EpEX: 9 nM EGF in combination with 50 nM EpEX-Fc. Scalebars represent 100 µm. F Principal component analysis of 3´-RNA-seq samples in Kyse30 and FaDu cells with the indicated treatments are shown (n = 4)