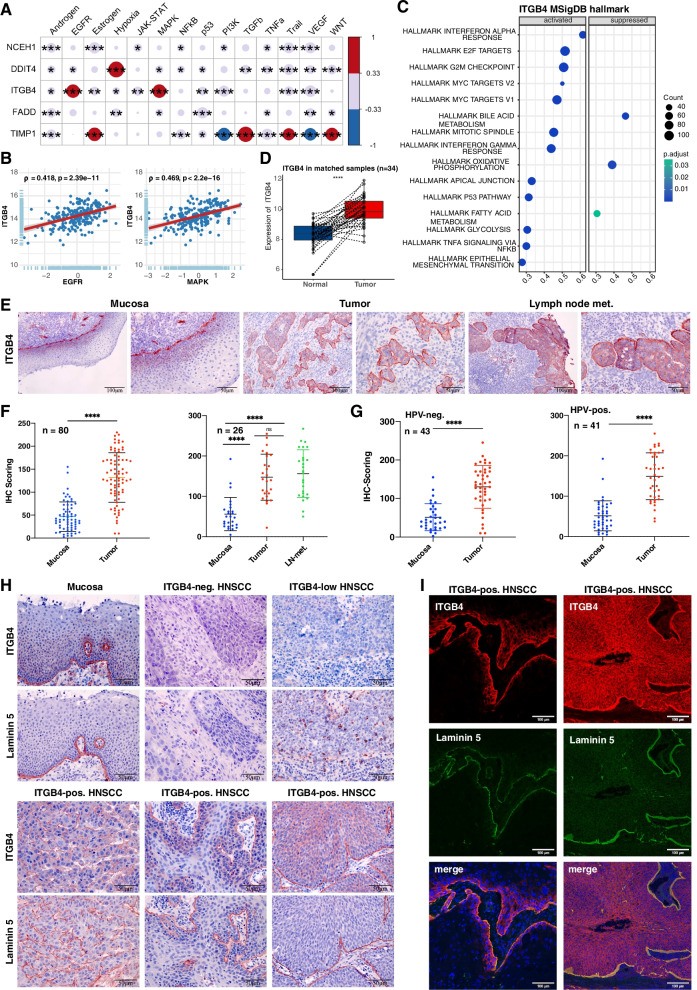
Fig. 5.
Integrin beta 4 (ITGB4) expression in HNSCC. A Pathway activities were inferred in HPV-negative TCGA-HNSCC samples using PROGENy (n = 240). Spearman correlations and p-values of the 5-gene signature with pathway activities are depicted. * ≤ 0.05; ** ≤ 0.01; *** ≤ 0.001. B Scatter plots of ITGB4 correlations with EGFR and MAPK pathways in HPV-negative HNSCC of TCGA patients (n = 240) are shown with Spearman correlation and p-value. C ITGB4-correlated genes were subjected to a GSEA using MSigDB hallmark gene sets. Top 15 regulated hallmarks are depicted with gene counts and adjusted p-values. D ITGB4 expression was compared in matched pairs of normal mucosa and HPV-negative HNSCCs of TCGA (n = 34). Matched expression values are shown in a box plot whiskers graph (t-test **** ≤ 0.0001). E Representative examples of ITGB4 expression in normal mucosa (n = 64), primary tumor (n = 80) belonging to n = 84 patients, and in triplets including lymph node metastases (n = 26) from HPV-neg. and -pos. HNSCC of the LMU cohort. F-G IHC scoring of ITGB4 protein expression is shown in scatter dot plots with means and SD for all samples and stratified according to HPV-status. Ns: not significant, **** ≤ 0.0001 (t-test and One-way ANOVA). H Representative examples of ITGB4 and laminin 5 expression in consecutive sections of normal mucosa and HNSCC are shown. I Double immunofluorescence staining of ITGB4 (red), laminin 5 (green), and DAPI (blue) in HNSCC with edge-localized (left) or homogeneous ITGB4 expression (right)