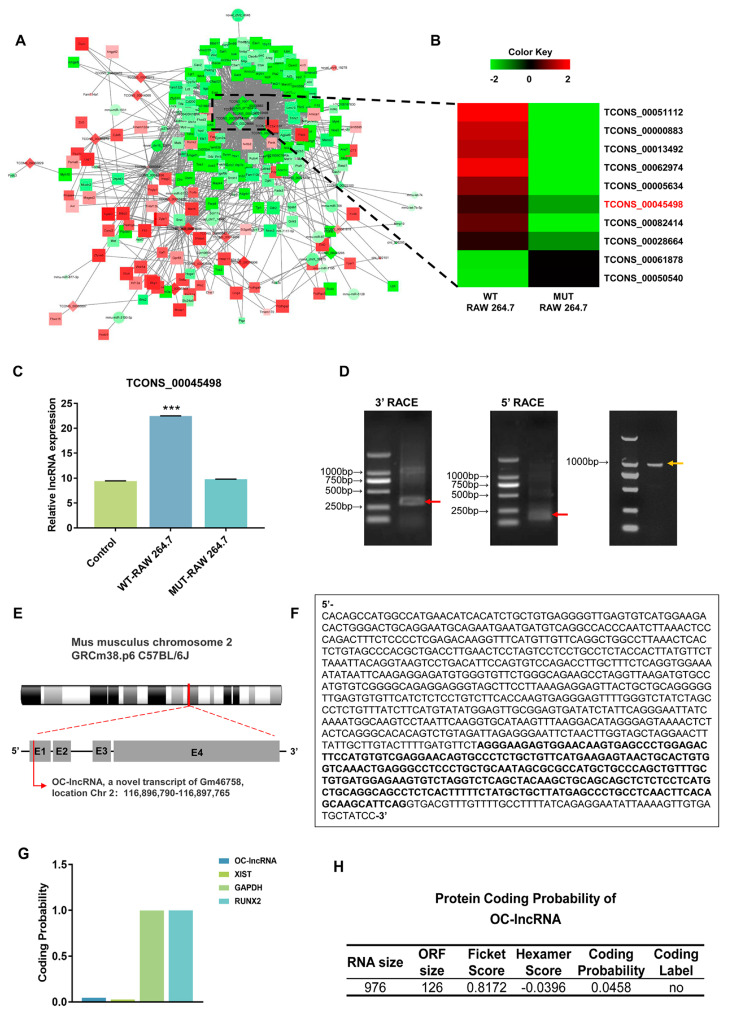
Figure 2.
Identification of a novel lncRNA transcript in CCD. (A) The regulatory network of differentially expressed mRNAs (rectangles), lncRNAs (rhombuses), microRNAs (circles), and circRNAs (triangles). (B) The heatmap of 10 core regulatory lncRNAs. (C) Expression validation of TCONS_00045498 by real-time PCR with RANKL induction for 3 days. (D) The gel results of the 3′/5′ RACE products (red arrows) and the whole sequence (yellow arrow). (E) Schematic of OC-lncRNA. (F) The complete nucleic acid sequence of OC-lncRNA. Identical sequence among OC-lncRNA and three transcripts of Gm46758 is indicated in bold. (G) Coding-potential assessment of OC-lncRNA by CPAT analysis. (H) OC-lncRNA displayed no coding probability. LncRNA XIST was used as a representative non-coding gene. GAPDH and RUNX2 were used as representative coding genes. Error bars represent SD. *** p < 0.001.