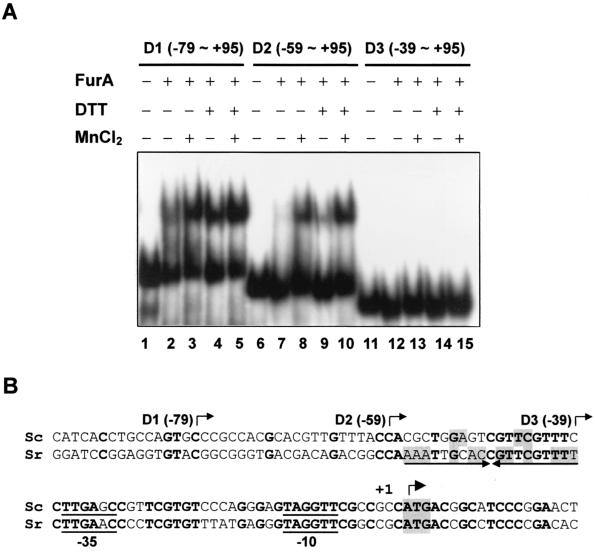
FIG. 7.
Mapping of the FurA binding site within the furA-catC promoter. (A) Gel shift assay for FurA binding. End-labeled DNA fragments (D1 to D3) containing the indicated region of the furA-catC promoter were analyzed for binding with partially purified FurA, which had been preincubated with (+) or without (−) 100 mM DTT. DNA and the FurA mixture were incubated at 30°C for 10 min in the presence or absence of 100 μM MnCl2 as indicated. The final concentration of DTT in the binding mixture was 10 mM. (B) Putative binding site of FurA. The nucleotide sequence alignment of the S. coelicolor (Sc) furA promoter region with the corresponding region of S. reticuli (Sr) furS is presented. The 5′ boundaries of the furA promoter fragments (D1, D2, and D3) used for gel shift assay are indicated by bent arrows. The inverted repeat sequence prominent in the furS gene is marked by arrows. The nucleotides identical between the two species are in boldface type. The inverted repeat nucleotides within the binding site are shaded. The translational start codon, ATG, is also shaded, and the transcription start site (+1) is indicated with another bent arrow.