Abstract
Plastics are inexpensive and widely used organic polymers, but their high durability hinders biodegradation. Polystyrene, including extruded polystyrene (also known as styrofoam), is among the most commonly produced plastics worldwide and is recalcitrant to microbial degradation. In this study, we assessed changes in the gut microbiome of superworms (Zophobas morio) reared on bran, polystyrene or under starvation conditions over a 3 weeks period. Superworms on all diets were able to complete their life cycle to pupae and imago, although superworms reared on polystyrene had minimal weight gains, resulting in lower pupation rates compared to bran reared worms. The change in microbial gut communities from baseline differed considerably between diet groups, with polystyrene and starvation groups characterized by a loss of microbial diversity and the presence of opportunistic pathogens. Inferred microbial functions enriched in the polystyrene group included transposon movements, membrane restructuring and adaptations to oxidative stress. We detected several encoded enzymes with reported polystyrene and styrene degradation abilities, supporting previous reports of polystyrene-degrading bacteria in the superworm gut. By recovering metagenome-assembled genomes (MAGs) we linked phylogeny and functions and identified genera including Pseudomonas , Rhodococcus and Corynebacterium that possess genes associated with polystyrene degradation. In conclusion, our results provide the first metagenomic insights into the metabolic pathways used by the gut microbiome of superworms to degrade polystyrene. Our results also confirm that superworms can survive on polystyrene feed, but this diet has considerable negative impacts on host gut microbiome diversity and health.
Keywords: polystyrene, plastic, degradation, microbial, bacteria, styrofoam, worm, metagenomics
Data Summary
All sequencing data have been deposited with the National Center for Biotechnology Information (NCBI) under BioProject accession number PRJNA801070. Supplementary material can be found on Zenodo: https://zenodo.org/record/6519352.
Impact Statement.
Increasing plastic pollution is a major environmental problem, and a recently proposed way to counteract this trend is to embrace a circular economy, in which used materials are recycled, rather than disposed of. An important step to facilitate this process is to invent new approaches for upcycling of plastic waste to desirable consumer products. Microbial plastic degradation and conversion is likely to play a considerable part in shaping a circular economy, by engineering microbes or their enzymes to bio-upcycle plastic waste. A first step towards actualizing this goal is to identify microbes that can degrade polystyrene and to investigate the enzymes and pathways involved. Our study represents the first metagenomic analysis of an insect gut microbiome on a polystyrene diet. It identifies bacteria with polystyrene- and styrene-degrading abilities, and infers enzymes and pathways involved in these reactions. Therefore, our results contribute towards understanding microbial polystyrene degradation and will provide a base for future investigations into microbial upcycling of plastic waste.
Introduction
Plastics are integral to the global economy and have become a part of our daily lives by providing excessive amounts of short-lived and disposable items [1]. Global plastic production reached nearly 360 million tonnes in 2018 and plastic demand is predicted to grow substantially over the next decade, while recycling rates are likely to remain low [2]. This lack of recycling combined with the high durability of plastics has resulted in a wide range of negative environmental impacts [3]. One of the most prevalent polymers globally is the durable thermoplastic polystyrene (PS), which accounts for up to 7–10% of the total non-fibre plastic production [4, 5]. Among the various PS types, PS foam is most widely used in consumer products [6] and is commonly referred to as styrofoam. Like other plastics, PS persists in nature for decades [7, 8], and can potentially harm wildlife and subsequently humans [3, 9]. Historically, PS was considered to be recalcitrant to microbes [10], and studies of PS degradation focused mainly on chemical and mechanistic forces [11, 12]. Indeed, the high molecular weight and hydrophobic character of PS polymers make them a difficult target for biodegradation since hydrophobic polymers are less susceptible to hydrolysis, which is the main pathway of natural polymer degradation [13]. However, initial abiotic degradation of PS has been reported to increase the polymer’s bioavailability in the environment by creating more exposed surfaces for biodegradation [12, 14, 15]. Over recent decades, several microbial isolates have been reported to utilize PS as a carbon source, albeit at a slow pace, with a reported PS weight loss ranging from 0.13 to 7% per day (Table S1, available in the online version of this article). Thereby, environmental PS biodegradation is probably enhanced by cooperative processes involving microbial consortia. Indeed, mixed communities from soils and liquid enrichment cultures have been reported to degrade PS [16], and marine microbial consortia effectively reduced the weight of PS films, suggesting that they are capable of degrading weathered PS pieces [17].
Insect gut microbiomes hold promising potential for efficient plastic degradation by combining both physical and biochemical mechanisms. The insect uses mechanical force, i.e. its oral appendages, to physically break up and ingest plastic particles which can subsequently be further degraded by the microbial community in the host’s digestive tract. Indeed, several recent studies have reported that insect microbiomes are capable of plastic degradation. Two bacterial strains, Enterobacter asburiae YT1 and Bacillus sp. YP1, isolated from the gut of waxworms, a common name for the larvae of the Indian mealmoth Plodia interpunctella, were capable of degrading polyethylene (PE) [18]. Larvae of the wax moth, Galleria mellonella, have also been reported to digest PE, yet the role of its intestinal microbiome is not well understood [19], and the reported evidence for PE biodegradation, in particular the detection of ethylene glycol, has been called into question [20].
Currently, the best characterized plastic-degrading insect gut microbiome is that of mealworms, a common name for the larvae of the mealworm beetle Tenebrio molitor (Tenebrionidae) [21, 22]. Mealworms were able to survive on PS as a sole diet, and degraded long-chain PS molecules in their gut, resulting in the formation of depolymerized metabolites [22]. The majority of PS was converted into CO2 (47.7%) and egested as faeces (49.2%), with only a limited amount (0.5%) being incorporated into biomass. The gut microbiome was deemed essential for the reported PS breakdown as mealworms, treated with antibiotics, lost the ability to depolymerize PS [21]. Furthermore, the Firmicute Exiguobacterium sp. strain YT2 isolated from a mealworm gut successfully formed a biofilm on PS and was able to degrade 7.4±0.4% of the PS pieces over a 60 day incubation period [21]. Another study, investigating the degradation of PE and mixed plastics in T. molitor, reported comparable biodegradation rates for PS and recovered two operational taxonomic units (OTUs) ( Citrobacter sp. and Kosakonia sp.) associated with both PE and PS degradation [23]. The authors concluded that, based on the observed degradation of mixed polymers, plastic degradation within the mealworm gut is non-specific in terms of plastic type.
Superworms, a common name for the larval stages of the darkling beetle Zophobas morio Fabricius, 1776 (Coleoptera, Tenebrionidae), have also been reported to survive on PS as the sole diet [24, 25]. Recently, the larvae were found to convert 36.7% of the ingested PS carbon to CO2 in a study using the heterotypic synonym Zophobas atratus Kraatz, 1880 of this species [25]. As in the mealworms T. molitor, the PS-degrading capabilities of superworms were inhibited by antibiotic suppression of gut microbiota, emphasizing the importance of the gut microbiome in PS degradation [25]. However, a metagenomic analysis of the superworm gut microbiome, identifying key community members and their inferred functions, is currently missing.
This study is the first metagenomic analysis of a plastic-associated insect microbiome. We aimed to confirm the survival of superworms reared solely on PS, and to investigate changes in the superworm’s gut microbiome in response to this diet. In particular, we compared differences in community compositions and inferred metabolic functions of the gut microbiome from superworms on a standard bran diet, a sole PS diet and under starvation conditions. We found >95% survival rates among superworms and a marginal weight gain in the PS group compared to starvation conditions. We identified the main microbial players in the gut microbiome during each diet regime, successfully reconstructed microbial genomes and inferred metabolic functions, including enzymes linked to polystyrene and styrene degradation.
Methods
Superworms and polystyrene
Superworms, the larvae of Zophobas morio, ranging in size from 25 to 40 mm, and superworm pollard (bran) used in this study were purchased from Livefoods Unlimited. Styrofoam was originally a trademarked brand of a light-blue extruded polystyrene (XPS) foam used in building insulation and has since become a generic name for white expanded polystyrene foam (EPA) frequently used in food containers, coffee cups and packaging material. The styrofoam used in this study was EPA purchased from Polystyrene Products (https://www.polystyreneproducts.com.au/). According to the manufacturer, the styrofoam contained at least 92–95% PS (CAS No. 9003‐53‐6), 4–7% pentane (CAS No. 109‐66‐0), an expanding agent added to PS beads and 1% of the flame retardant hexabromocyclododecane (CAS No. 25637‐99‐4) at the time of manufacture. The styrofoam was stored in our laboratory over several months prior to the feeding trials and was therefore considered to contain only trace amounts of pentane, since this agent diffuses to insignificant levels within several weeks after production [26].
Feeding trials
Initial trials with groups of starving superworms confirmed reports of cannibalism [27], which led to our modified experimental design housing the starving control group animals in isolation, whereas animals in the other two groups were housed together during the feeding trial (Fig. S1). While isolation might influence social behaviour and could potentially impact the microbiome, we found that this setup is the only effective way to prevent cannibalism under starvation conditions.
A total of 171 superworms were housed in one container for the initial 24 h following arrival. Subsequently, the 171 superworms were split into three groups with three replicates of 15 superworms each, all of which were fed with wheat bran (certified organic wheat pollard; product code: MWORMPOL; Livefoods Unlimited) supplemented with carrots, during an initial 1 week of acclimatization period (Fig. S1). Moisture was controlled with a custom water sprayer to create a fine mist layer on the underside of the superworm container lid. All experiments were carried out at room temperature, which ranged from 20 to 25 °C during the experiment. After the acclimatization period, four superworms from all three replicates in each group (4×3×3=36) were flash-frozen in liquid nitrogen and stored at −80 °C. The remaining 135 superworms underwent the subsequent 3 week feeding trial (Fig. S1). Thereby each group, consisting of three replicates with 15 superworms each, received a different feed: (i) the bran group was fed solely with wheat bran, (ii) the polystyrene (PS) group was supplied solely with untreated PS as a food source and (iii) the starvation control group received no food supply (Fig. S1). The weight of the superworms and all styrofoam blocks was recorded twice a week. After the 3 week feeding trial, up to 15 superworms from each group were separated for the pupation trial, and the remaining superworms were flash-frozen in liquid nitrogen and stored at −80 °C for metagenomics. For the pupation trial, the superworms were housed individually in separate containers (Fig. S1), since the larvae are known to fail to pupate under crowded conditions [28]. No diet was provided during the pupation trial and the results (pupae, live imago) were recorded over the following 4 weeks.
Superworm weight statistical analysis
Differences in superworm weight changes between groups were assessed by one-way ANOVA followed by Tukey’s honest significant difference (HSD) post-hoc test.
Dissection of the superworms and faeces collection
For dissections, the superworms were removed from the −80 °C storage, placed on their side under a dissection microscope and, while still frozen, cut along the lateral axis from behind the legs to the last abdominal segment. The larvae’s digestive tract was pulled out and a section including midgut and hindgut was removed for DNA extraction. Superworm faeces were harvested daily over a 1 week period, in order to accumulate enough material for sequencing analysis, and were stored at room temperature during that time. After the last collection on day 7 the faeces was stored at −80 °C until DNA extraction.
DNA extraction and sequencing
The Power Biofilm DNA Isolation Kit (Qiagen) was used for all microbial DNA extractions from the frozen samples without any pretreatment. In total, 15 samples were processed, which included 12 superworm gut tissues (three from the acclimatization experiment, and three from each of the three feeding trial groups), a faeces sample from the bran group, a faeces sample from the PS group as well as one negative control (200 µl water). Genome sequencing was carried out on the Illumina NextSeq 500 platform using Nextera XT libraries, which were created following the standard Illumina protocol with 12 cycles of limited cycle PCR amplification, with the only modification of reducing the reagent volumes to 1/5th of the tagmentation and the PCR step. The libraries were sequenced with a 2×150 bp high-output v2 run chemistry, and a targeted sequence allocation of 3 Gb per sample.
Read QC
Quality control of raw Illumina reads (2×150 bp) from all 15 samples was performed with FastQC cersion 0.11.9 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
16s rRNA/SSU rRNA gene analysis
We characterized the microbial community composition based on small subunit (SSU) rRNA gene abundances with GraftM, a tool that uses gene-specific packages to rapidly identify gene sequences followed by a taxonomic classification derived from placements into a pre-constructed gene tree [29]. In brief, SSU rRNA reads were detected and classified using the GraftM package ‘7.71.silva_v132_alpha1.gpkg’ which uses SSU rRNA sequences and the taxonomy-decorated phylogenetic tree based on the 99% nucleotide identity representative OTU set from the silva SSU database release 132, Ref NR 99 (https://www.arb-silva.de/). Subsequently, GraftM results were manually curated by removing lineages identified as contamination. In brief, sample contamination is a combination of crosstalk from nearby wells and background contaminants [30], and we therefore followed an approach outlined previously [31, 32], which is based on the presumption that contaminant taxa are expected to have high relative abundances in negative controls but low relative abundances in samples. Therefore, lineages that exhibited a relative abundance of one or more orders of magnitude higher in the negative control compared to any sample were removed. This resulted in the removal of 11 lineages including Cutibacterium , Comamonas and Marinococcus , as well as reads assigned to mitochondria and chloroplasts (Table S2). We further evaluated the proportion of eukaryotic host sequences in our samples with graftM (-euk_check) using the package ‘4.40.2013_08_greengenes_97_OTUs_with_euks.gpkg’.
Microbial diversity estimates
The Shannon diversity index and the Simpson diversity index were calculated. To negate potential rarefaction biases, we additionally randomly subsampled 500 times and then calculated the average Shannon index.
Assemblies
We performed genome assembly individually for each sample, as well as combined genome assembly for the three replicates from the same experimental condition using MegaHit v1.1.2 [33]. The individual assemblies were used for gene calling and annotations, whereas the combined assemblies were used for binning.
Bacterial gene annotation of assemblies – gene-centric analysis
Gene calling of metagenomic assemblies was performed by Prodigal [34]. Genes of metagenome assemblies were then blasted against the Uniprot TrEMBL database (Fig. S2) using Diamond’s blastp function [35]. Genes with a bacterial blast top hit were selected and annotated by KOfamScan [36] for the gene profiling analysis, while eukaryotic genes and genes without blast hits were excluded from the downstream analysis (Fig. S2). Gene calling and annotation was performed with enrichM (https://github.com/geronimp/enrichM), which uses HMMS to assign EC, KO, PFAM and TIGRFAM identifiers, and dbCAN to annotate CAZY. To cover assignments not included in the enrichM database, KO IDs were annotated by blasting against Uniprot using Diamond, whereas PFAM and TIGRFAM were searched against their respective databases using hmmsearch.
For the functional comparisons, replicates BR3, PS1 and CO1 were excluded from the analysis due to the low gene counts of bacterial genes.
To screen for differentially abundant genes, counts of genes with a bacterial blast top hit and a KOfamScan annotation (see above) were normalized and transformed with variance-stabilized transformation using DESeq2 in R (https://www.rdocumentation.org/packages/DESeq2/versions/1.12.3) from which a principal component plot (PCA) was created to determine the variation between the communities as well as differential abundances for the top 50 genes. We compared this approach to an alternative method to normalize our data, based on the trimmed mean of M-values (TMM) [37], using the edgeR package in Bioconductor [38].
To detect enriched KEGG modules, genes with assigned KEGG Orthologs (KO) IDs were rarefied to 8255 genes, based on the lowest number of genes present in one of our samples, and analysed with EnrichM classify, which assigns modules based on a matrix of KO annotations (https://github.com/geronimp/enrichM).
In silico PCR amplification
The primers (FW 5′-CGCCAGTTGCTCTGCCATCG-3′, RW 5′-TGCCATGTGG GCGACGCGGC-3′) designed to amplify serine hydrolase (SH) from Pseudomonas sp. DSM 50071 [39] were used for an in silico PCR amplification against Pseudomonas aeruginosa on the website http://insilico.ehu.es/PCR/ [40]. The resulting 300 bp nucleotide sequence was then blasted (blastX) to obtain SH protein amino acid sequences.
Recovery of metagenome-assembled genomes
Read mapping was performed with BamM v1.7.3 (http://ecogenomics.github.io/BamM/). UniteM (https://github.com/dparks1134/UniteM) and CheckM [41] were used for binning and bin quality control, respectively, followed by phylogenetic tree reconstruction and taxonomy assignment performed by gtdb-tk [42]. Functional analysis was performed with EnrichM (https://github.com/geronimp/enrichM) and KofamScan [36] as described for the ‘Bacterial gene annotation of assemblies’ above, but without excluding any genes from the analysis.
Phylogenetic inferences
Phylogenetic trees for the genes of SH, StyA and StyE were inferred from MAFFT- [43] aligned protein sequences using IQ-TREE 2 [44] with the settings ‘LG+C10+F+G+PMSF’ and a starting tree inferred with FastTree 2 [45].
Phylogenetic trees for metagenome-assembled genomes (MAGs) recovered in this study were inferred based on 122 protein markers aligned with gtdb-tk [42] using IQ-TREE 2 [44] with the settings ‘LG+C10+F+G+PMSF’ and a starting tree inferred with FastTree 2 [45].
Results and discussion
Host survival, behaviour and life cycle
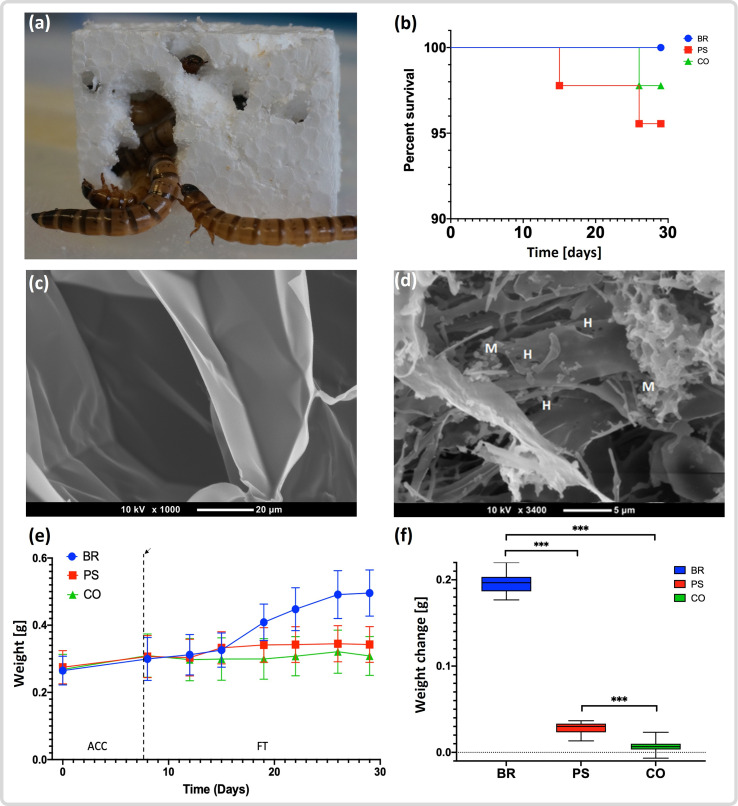
Following an initial 1 week acclimatization period, we conducted a 3 week feeding trial consisting of superworms from three groups; bran-reared, PS-reared and a starvation group (Fig. S1). Superworm survival rates in all three groups were above 95% over the entire duration of the feeding trial (Fig. 1a). This is in contrast to a previously reported survival rate of ~70% after 3 weeks, conducted under similar experimental settings but with over 10 times higher superworm densities [25]. It is likely that our experimental design prolonged survival by avoiding overcrowding, which can lead to aggressive behaviour and fatalities among worms, as reported for the common mealworm [46]. Superworms in the PS group readily approached the PS blocks and created narrow burrows by chewing their way into the blocks within the first 24 h of the experiment (Fig. 1b). While we did not quantify superworm activities, we made several anecdotal observations. Worms subjected to the PS diet remained active during the duration of the experiment, although they moved about with a slower speed than worms in the bran group. The starvation group without feed showed the least movement, with prolonged resting periods interspersed by short explorations.
Fig. 1.
Superworm polystyrene utilization, survival rates and weight changes. (a) A group of superworms eating their way into a polystyrene block. (b) Superworm survival rates for the three groups: bran (BR, blue), polystyrene (PR, red) and the starvation control group (CO, green) during the duration of the 3 week feeding trial. (c) Scanning electron micrograph of the extruded polystyrene (styrofoam) as supplied by the manufacturer. (d) Polystyrene particle recovered from the gut of a superworm in the PS group. Note several holes (H), and microbial cells (M). (e) Weight change of the superworms in the three groups. Day 1 to day 7 represents the acclimatization period (ACC) and the end of the ACC is indicated by a dashed line. (f) Overall weight change of the superworms in each group at the end of the feeding trial. Asterisks indicate that the weight change between BR and PS, BR and CO, and PS and CO was significantly different (Table S3). Box plots show the median (bold line), first and third quartile (box outline), 10th and 90th percentile (lower and upper whisker boundaries), and outliers (circles).
The faeces changed colour from light brown to white pellets in the first 24–48 h in the PS group, suggesting that the worms had started to consume and egest PS. Dissections of the worm’s digestive tracts revealed that the midgut and hindgut of individuals in the PS group were tightly packed with PS particles (Fig. S3). Electron microscopy confirmed that, compared to virgin extruded PS, the PS particles in the superworm guts appeared partially degraded, and showed attached microbial cells (Figs 1c, b and S4). We therefore conclude that superworms were able to ingest PS and pass it through their entire gastrointestinal system, where it came into contact with the gut microbiome.
The average weight of superworms in the bran group more than doubled during the experiment, leading to significantly heavier worms (P<0.001) compared to the PS and the starvation group (Fig. 1c and d). The average superworm weight in the PS group increased slightly during the feeding trial (Fig. 1c), resulting in a marginal but significant weight gain (P<0.001) at the end of the feeding trial (Fig. 1d, Table S3). This weight gain and the anecdotal observations of higher activity compared to the starvation group suggest that worms in the PS group were able to obtain energy from the PS feed, probably in cooperation with their microbial gut communities. This hypothesis is supported by previously reported carbon recovery efficiencies, which indicated that mineralization of ingested PS occurs in the superworm gut [25].
To assess the impact of the PS diet on the lifecycle of Zophobas morio, we conducted a pupation trial following the 3 week feeding experiment (Fig. S1). We limited the trial to 10–15 worms from each group and observed a pupation rate of 92.9, 66.7 and 10.0% for the three groups, bran, PS and starvation, respectively (Fig. S5). All formed pupae completed the entire pupation phase and emerged as adult beetles. We hypothesize that the marginal weight advantage of worms in the PS group increased the likelihood of a successful pupation nearly seven-fold compared to the starvation group. Weight gain in insect larvae is mainly due to an increase in fat reserves [47], and therefore the lower body weight in the control group could have resulted in insufficient fat reserves prohibiting a complete metamorphosis. However, additional experiments are needed to verify this hypothesis. Next, we analysed the microbial gut community of the superworms, to identify microbes, encoded pathways and enzymes potentially involved in PS degradation.
Microbial community composition
Mid- and hindguts of superworms (Fig. S3) were dissected and, together with two faecal samples, used for DNA extraction and metagenomic sequencing (see Methods). The obtained shotgun reads were mined for SSU rRNA gene sequences to omit insect host genes (Fig. S6) and to characterize prokaryotic microbial gut communities. On average, the microbial diversity decreased from bran (H′=2.66±0.29) to PS (H′=2.54±0.19) and further from PS to starvation conditions (H′=2.24±0.15) (Table S4), suggesting that PS is a poorer diet compared to bran, but still supports a more diverse community compared to starvation conditions. This conclusion aligns with previous reports demonstrating that microbiome diversity is positively correlated with diet diversity [48], a trend that has also been confirmed for insect larvae [49, 50]. The lowest microbial diversity, observed in the starvation group, indicates a lack of nutrients or even the onset of a disease. The latter assumption is supported by the detection of pathogenic bacteria (see below) and reports that a loss of gastrointestinal species diversity is a common finding in several vertebrate disease states [48]. This correlation might translate to insect gut microbiomes, since greater microbial diversity has been associated with health benefits for insect hosts [51].
Overall, the gut microbiome of the baseline acclimation, bran, PS and starvation group was dominated by the two bacterial phyla, Firmicutes and Proteobacteria with maximum relative abundances of 80.6 and 67.8%, respectively (Fig. 2, Tables S5 and S6). Other phyla, including Tenericutes , Bacteroidetes and Actinobacteria , were also present, although at lower relative abundances with a maximum of 15.9, 15.1 and 11.5%, respectively (Table S6, Fig. 2). This observed superworm microbial gut community is similar to microbiomes from common mealworms (T. molitor), with studies reporting Firmicutes , Proteobacteria but also Tenericutes and Actinobacteria as dominant phyla [52, 53]. Interestingly, no archaeal SSU rRNA sequences were detected in our superworm guts, similar to findings from a previous analysis of mealworms [54]. This probably indicates that Archaea are only present in very low abundances below detection limits or are not part of mealworm and superworm gut microbiomes.
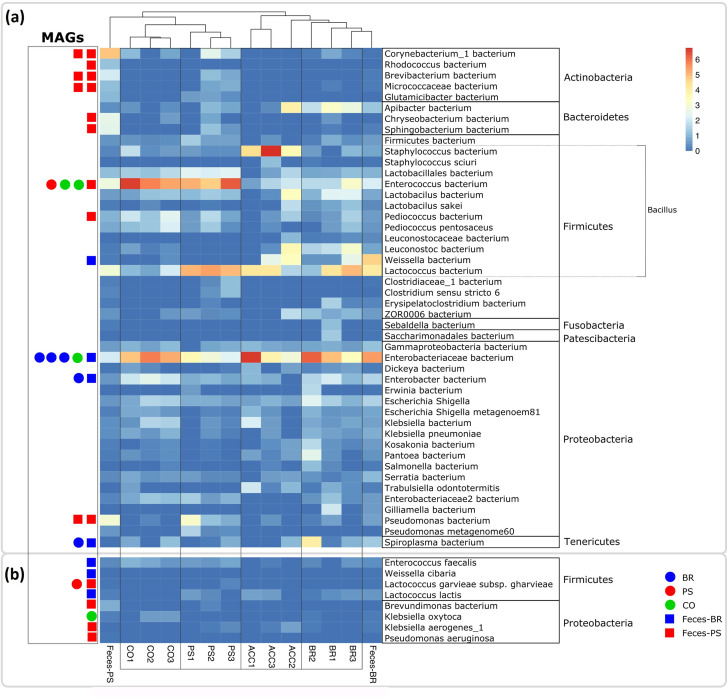
Fig. 2.
Abundant bacterial taxa in the superworm gut microbiome. The community profile is based on SSU rRNA short read alignments against the Silva r132_99 database. (a) Taxonomic lineages with a relative abundance >1 % were included in the heatmap. Abbreviations: CO1–3 (replicates of the starvation control group without feed), PS1–3 (replicates of the polystyrene group), ACC1–3 (replicates of the acclimation baseline group sampled before the feeding trial), BR1–3 (replicates of the wheat bran group), Feces-PS (polystyrene faeces), Feces-BR (bran faeces). MAGs recovered from our samples are indicated with a circle (feeding groups) or a square (polystyrene and bran faeces samples) next to the assigned lineages. (b) Eight lineages with a relative abundance <1 % from which we recovered MAGs. The highest assigned taxonomic rank is provided for each lineage, and phylum assignments are indicated on the right. In addition, within Firmicutes the class Bacillus is indicated by a dashed line. Bar, square root-transformed relative abundances from the lowest (zero) to the highest (>6) value.
Among the most abundant taxa in the bran group were facultative anaerobes from the Gammaproteobacteria family Enterobacteriaceae , including the genera Enterobacter, Escherichia/ Shigella , Erwinia , Klebsiella and Dickeya (Fig. 2). These taxa have been reported from insect guts previously [55] and some lineages including Enterobacter , Klebsiella and Erwinia are known for mutualistic relationships with their insect hosts [55–58]. Other genera such as Dickeya contain mainly plant pathogens [59] with a potential to kill insect hosts [60]. Overall, the bran microbiome did not change significantly compared to the baseline group (Fig. 2), suggesting that the worms became well acclimatized before the feeding trials commenced. The only exception was an uncultured species in the genus Staphylococcus which showed significantly higher relative abundance in the acclimation group compared to all other groups (Figs 2 and 3a–c). Further abundant taxa in the bran group were lactic acid bacteria from the class Bacilli including the genera Lactoccocus, Weisella, and Leuconostoc , which contain species with reported probiotic effects [61, 62]. In particular, Weisella, Leuconostoc and Lactoccocus were significantly enriched in the bran group compared to the PS and starvation group, respectively (Fig. 3d, e), indicating that these taxa might benefit from a fibre- and nutrient-rich bran diet.
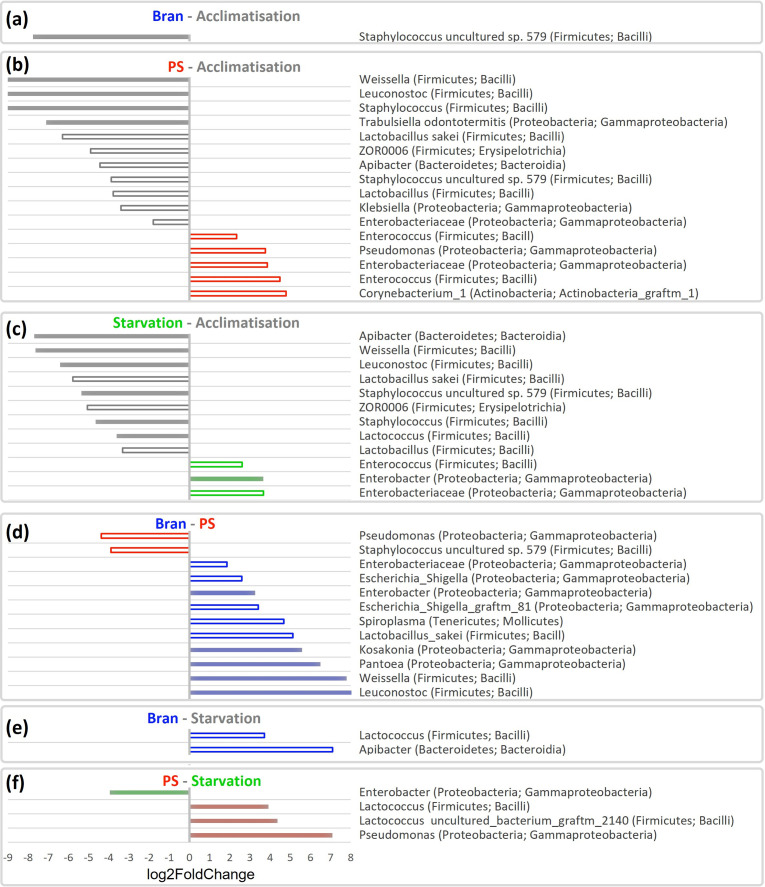
Fig. 3.
Gut microbiome lineages with significant differences in abundances between feeding trial groups. Estimated log2fold changes with an adjusted P-value of <0.1 (colour filled bars) and <0.05 (white filled bars) comparing the feeding trials: (a) bran vs acclimatization, (b) polystyrene vs acclimatization, (c) starvation vs acclimatization, (d) bran vs polystyrene, (e) bran vs starvation, and (f) polystyrene vs starvation. The highest assigned taxonomic rank is provided for each lineage and phylum and class assignments are given in parentheses. PS, polystyrene. Colour code: bran (blue), polystyrene (red) and the starvation control group (green). Note that the log2fold change value indicates how much the abundance has changed between the comparison and reference groups, and that each panel is labelled with the comparison group, followed by a dash and the control group (e.g. ‘Bran - PS’ in d). Here, a positive log2fold change value indicates that a taxon has a significantly higher abundance in the comparison group, and a negative log2fold change value means a taxon has a significantly higher abundance in the reference group.
The microbiome of the PS group superworms differed from the bran group by a lower relative abundance of the family Enterobacteriaceae (Fig. 2). In particular, the Enterobacteriaceae genera Kosakonia , Pantoea , Weisella, Leuconostoc and Enterobacter had significantly lower abundances compared to the bran group (Fig. 3d). The PS group was further characterized by higher relative abundances of the Bacilli genera Lactococcus and Enterococcus , whereby the latter genus was also elevated in the starvation group (Fig. 2). The presence of Enterococcus, a genus of facultative anaerobes [63], in the superworm gut is not surprising since several Enterococcus species were isolated from gastrointestinal tracts of animals, including a variety of insects [64]. Members of this genus have also been associated with plastic substrates. Multiple Enterococcus species were found to survive over 90 days on polyester and polyethylene fabrics in hospitals [65], and elevated levels of Enterococcus avium were reported from mealworms reared on a diet of extruded PS [54]. Our data confirm the association of E. avium with a PS diet, since we detected this species in the PS but not in the bran and starvation groups (Table S7), although with low relative abundances (<0.2 %). However, given the large phylogenetic and metabolic diversity of the genus Enterococcus [63], it is possible that some taxa act as pathobionts in the insect gut, similar to reports from mammals [66].
In addition to superworm gut samples, we analysed a faecal sample from the PS and the bran group, respectively. While the bran faecal sample clustered with the bran group, the PS faecal sample was distinct from all other samples. The dominating lineage was Corynebacterium , a genus that was also moderately abundant in the PS group (Fig. 2). The genus Corynebacterium comprises mostly aerobic bacteria associated with a range of host organisms, including humans [67]. Corynebacterium , as defined in NCBI, is divided into two genera in the silva database, named Corynebacterium and Corynebacterium_1. The latter genus is present in our samples (Fig. 2) and has been reported as an opportunistic pathogen linked to dysbiosis in the gut microbiome of patients with Parkinson’s disease [67].
The starvation group gut microbiome shared the high relative abundance of Enterococcus with the PS group but had a significantly lower level of Lactococcus and a significantly higher level of Enterobacter compared to the PS group (Figs 2 and 3). The elevated presence of the pathogen Klebsiella oxytoca (Fig. 2), an emerging human pathogen causing hospital-acquired infection [68], supports previous findings of an elevated K. oxytoca presence in starved insect larvae, based on experiments with the common mealworm T. molitor [54].
In conclusion, we hypothesize that higher relative abundances of Enterococcus in the PS and starvation groups, of K. oxytoca in the starvation group, and of Corynebacterium_1 in the PS faeces indicate that a PS diet and starvation conditions result in poor gut health and potentially induce dysbiosis in the superworm gut. Future studies combining microbial community profiling and gut histomorphology are necessary to test this hypothesis.
Microbial taxa associated with polystyrene degradation
Over the last two decades, a range of bacterial strains have been enriched or isolated with PS as the sole carbon source, and most were identified at the genus or species level (Table S8). Based on our SSU assignments, we detected six of these genera (Bacillus, Brevundimonas, Microbacterium, Pseudomonas, Sphingobacterium and Streptococcus ) with higher relative abundances in the PS group compared to all other groups (Table S8). In particular, Pseudomonas had significantly higher relative abundances in the PS group compared to the bran group (Fig. 3d). Various species from this genus have been previously isolated with PS as the sole carbon source (Table S1), and we detected two, Pseudomonas aeruginosa and P. putida, solely in the PS faeces, albeit with low relative abundances below 0.02% (Table S8). Further PS-degrading taxa, classified at the species level (Table S1), detected in our samples include Brevundimonas diminuta uniquely detected in the PS group, and Rhodococcus ruber exclusively detected in the PS faeces (Table S8). Interestingly, the genus Kosakonia , reported to have a strong association with PS and polyethylene diets in the common mealworm [23], had a significantly lower abundance in the PS group compared to the bran group (Fig. 3d). This suggests that different Kosakonia species or strains could be present in the gut of superworms and mealworms, respectively. Future deep metagenomic sequencing might be able to provide the necessary taxonomic resolution to resolve this question.
Functional community profiles
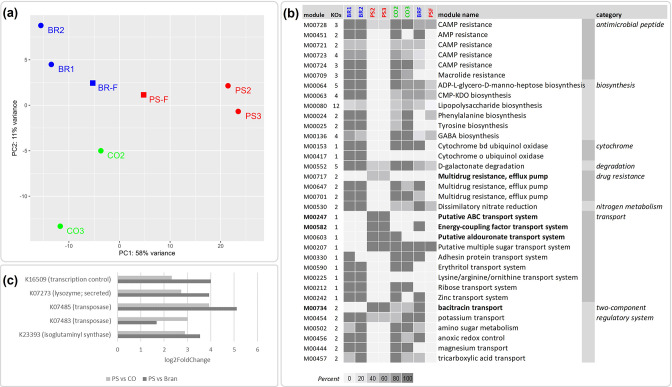
To assess changes in the functional potential of gut microbiomes across samples, we assigned genes to bacterial KO identifiers (Table S9). Overall, the encoded microbiome functions differed considerably among bacterial genes from the bran, PS and starvation groups, while clustering according to diet (Fig. 4a). The PS group was characterized by the absence or incompleteness of several biosynthesis modules, including amino acid, lipopolysaccharide and GABA biosynthesis, as well as a lack of antimicrobial peptides, drug resistance, transport and two-component regulatory systems (Fig. 4b). Modules enriched in the PS group include transporters such as the putative aldouronate transport system (Fig. 4b), which has been reported to facilitate the intracellular conversion of aldouronates following extracellular depolymerization of hemicellulose in bacteria [69]. However, whether these transporters aid in the import of depolymerized PS remains to be determined.
Fig. 4.
Functional profiles of superworm microbiomes. (a) PCA of genes assigned to bacterial KEGG Orthologs (KO identifiers), showing a clear separation of all three feeding trials, including bran (BR; blue dots), polystyrene (PS; red dots), starvation control group (CO; green dots), as well as polystyrene faeces (PSF; red squares) and bran faeces (BRF; blue squares). (b) KEGG modules enriched and depleted in the polystyrene (PS) group based on rarefied gene counts. Shown are the KEGG modules, the number of KOs per module and the module completeness (0–100 %) reported for the bran samples (BR1, BR2), polystyrene samples (PS2, PS3), and the starvation control samples (CO2, CO3). Modules present in the PS group but absent in the bran group are highlighted in bold. (c) Differential abundant genes, with KO identifiers, enriched in the PS group compared to the starvation control (CO) and bran group, respectively. Note that gene counts were rarefied to 8255 for the module comparison in (b).
A small subset of genes assigned to functional homologues were differentially more abundant in the PS group compared to the bran and the starvation group (Fig. 4c), indicating a possible association with a PS diet. Two genes were identified as bacterial transposases (Fig. 4c, Table S10). Transposases are known to facilitate gene duplications and genomic rearrangements in bacteria, and are considered to be means of genome adaptations to survive stressful situations following environmental changes [70]. The unbalanced diet in the PS group could represent such challenging conditions for the bacterial gut community, resulting in higher abundances of transposases as a strategy to facilitate rapid adaptation to cope with the stressful environment [71]. Genes enriched in the PS group also suggest that the PS microbiome has a greater potential for membrane restructuring. This conclusion is supported by enriched genes encoding the lysosome M1 (GH25 family, K07273), an enzyme involved in remodelling peptidoglycan and in cell lysis for the dissemination of phage progeny [72], and isoglutaminyl synthase (K23393, murT) involved in peptidoglycan biosynthesis [73]. Whether this increased membrane plasticity is another stress response needs to be determined. However, the transcriptional regulator protein spxA (K16509), which is responsible for transcriptional control during oxidative stress and may cause genome-wide changes in expression patterns [74], was also enriched in the PS group, indicating that increased oxygen exposure might be a stress-contributing factor. We screened for aerobic and anaerobic modules across all samples to investigate if the nearly anoxic conditions, which exist in insect guts [75], might have been influenced by the PS diet. Our results showed an increased presence of encoded aerobic functions in the PS group and the PS faeces, compared to all other samples (Table S11). This supports our conclusion of deteriorating gut conditions in the PS group (see ‘Microbial community composition’), as a healthy gut epithelium is expected to consume oxygen, generating a state of hypoxia in humans [76] and probably also in insects [77].
Furthermore, we hypothesized that a shift in diet from bran, i.e. wheat pollard that is a by-product of the flour milling of grain and has a high fibre content [78], to PS will lead to a fibre-deprived gut microbiome. Such a microbiome can degrade the colonic mucus barrier and enhance pathogen susceptibility, as shown in mouse models [79]. Indeed, a functional characterization of encoded carbohydrate active enzymes (CAZy) revealed differences between samples (Fig. S7) but did not detect enriched or depleted fibre or mucin targeting CAZy genes, probably due to the fact that such changes commonly occur on a transcriptional level [79].
Detection of potential PS- and styrene-degrading enzymes
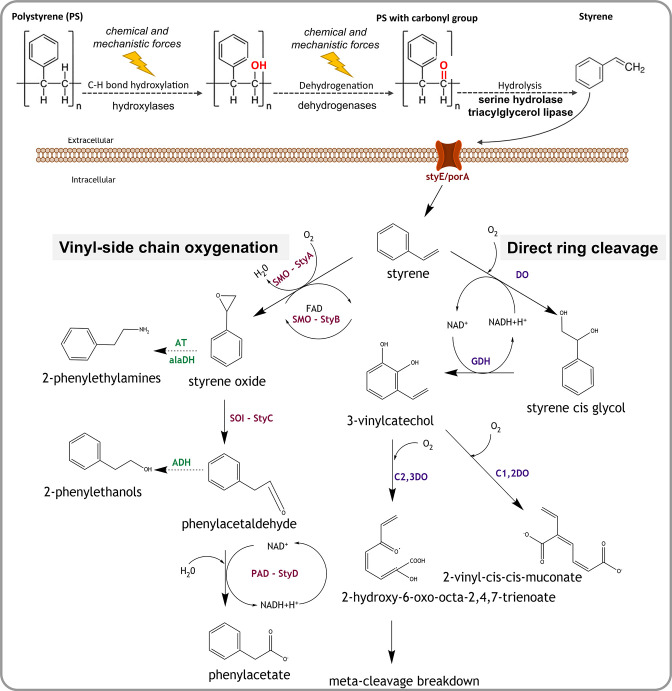
The lack of characterized enzymes for the initial step in microbial PS degradation, the breakdown of the insoluble PS polymer into styrene monomers, has led to the conclusion that the environmental degradation of this synthetic polymer depends initially on abiotic factors. UV irradiation and mechanical forces, such as wave action, wind, climate, or presumably the shredding and ingesting of PS by superworms, attack the PS backbone that contains only carbon–carbon bonds and has no ‘hydrolysable’ groups. This abiotic stress causes the polymer to age, that is, to react with small environmental molecules, typically O2 or H2O, resulting in the formation of carbonyl groups [80] which are considered more accessible to enzymatic degradation [10, 81, 82]. However, microbial pathways to age PS polymers seem plausible and could utilize hydroxylases for oxygen insertion [83] and subsequently dehydrogenases to form carbonyl groups (Fig. 5). This process could be catalysed by extracellular enzymes, similar to fungal extracellular cellobiose dehydrogenases [84]. Subsequently, enzymes with catalytic activities similar to phenolic acid decarboxylases [85], lipases [86], serine hydrolases [39] or the recently characterized polyethylene terephthalate (PET)-degrading enzyme PETase [87] could facilitate a further PS breakdown by targeting carbonyl groups. Currently, the best evidence for an enzymatic carbonyl group-based PS degradation comes from an analysis of the aforementioned serine hydrolase gene, reported from a Pseudomonas aeruginosa strain isolated from a superworm gut under oxygen-free conditions [39]. The authors report that quantitative PCR-based SH expression levels were elevated when growing the Pseudomonas strain on a nutrient-limited medium with added PS. Furthermore, PS degradation did not occur when the SH was blocked with an inhibitor, leading the authors to conclude that SH is involved in the breakdown of PS by targeting carbonyl groups, probably resulting in styrene dimers or monomers [39]. Since the authors did not sequence the reported SH gene [39], we performed an in silico PCR, followed by a blastp detection of this gene in our dataset. Among the feeding trials, we recovered SH homologues solely from the PS group, but both PS and bran faeces assemblies contained copies of this gene as well (Table S12). Phylogenetic inferences revealed that the SH genes recovered in our study clustered with the homologue from P. aeruginosa , indicating that the encoded superworm microbiome SH proteins can facilitate PS breakdown (Fig. S8). However, further culture-based experiments, including transcriptomics and gene knockout trials, are necessary to clarify the function of SH and other genes in the PS degradation pathway of this and related P. aeruginosa strains present in the superworm gut microbiome.
Fig. 5.
Potential pathways and enzymes of bacterial polystyrene and styrene degradation. Initially, abiotic chemical and physical factors, and potentially also extracellular enzymes such as hydroxylases and dehydrogenases, attack the polystyrene polymer creating carbonyl groups. Next, extracellular enzymes such as serine hydrolase and triacylglycerol lipase degrade the polymer to styrene monomers. A putative transmembrane protein StyE/PorA can import styrene monomers into the cell, where it is degraded via the vinyl side chain oxygenation or the direct ring cleavage pathway. In the vinyl side chain oxygenation pathway, styrene is broken down to phenylacetate using enzymes of the styABCD operon: styrene monooxygenase (StyA/SMO), styrene monooxygenase reductase component (StyB), styrene oxide isomerase (StyC/SOI) and phenylacetadehyde dehydrogenase (StyD/PAD/FeaB/ TynC). Styrene can also form 2-phenylethylamines and 2-phenylethanols as intermediates, through modifications of vinyl side chain pathway by the enzymes aminotransferase (AT), alanine dehydrogenase (alaDH) and alcohol dehydrogenase (ADH). The direct ring cleavage pathway converts styrene into Krebs cycle intermediates via meta cleavage breakdown or to muconic acid. Involved key enzymes are styrene dioxygenase (DO), 2,3-dihydrodiol dehydrogenase/cis glycol dehydrogenase (GDH), catechol-1,2-dioxygenase (C1,2DO) and catechol-2,3-dioxygenase (C2,3DO). Dashed arrows indicate hypothetical reactions.
Another group of hydrolases with potential function in polymer degradation are esterases. In particular, lipases, a subclass of esterases, have been experimentally verified to perform surface hydrolysis of synthetic polymers, including PET fabrics and films [88]. Furthermore, triacylglycerol lipase (EC 3.1.1.3), as well as two esterase carboxylesterases (EC 3.1.1.1) and cutinases (EC 3.1.1.74), have been identified as probable enzymes for the enzymatic hydrolysis of difficult to degrade aliphatic-co-aromatic polyesters [89]. An unspecified lipase has also been reported from a high impact polystyrene (HIPS) degrading Bacillus species [86]. Based on these data, we screened the genes recovered from all samples for lipase, carboxylesterase and cutinase homologues. The only esterase genes detected were homologues encoding triacylglycerol lipase, which we found exclusively in the PS group and in the PS faeces (Table S13). We therefore hypothesize that this lipase homologue could be involved in PS degradation by acting on the carbonyl group of the partially degraded PS, next to its traditional function of attacking the ester carbonyl of fatty acids. Furthermore, lipases are known to act on medium- to long-chain polymers with more than ten carbon atoms [90], which makes these enzymes potentially more suitable for the degradation of partially degraded synthetic polymers; however, experimental verification is needed to test this hypothesis.
Once PS is degraded to styrene, these monomers can be further broken down by a variety of microbial pathways, owing to the fact that styrene occurs naturally in plants and fungi [91]. Two main pathways have been described to facilitate this breakdown under aerobic conditions, direct ring cleavage and vinyl side chain oxygenation [92–94]. Initially, side chain oxygenation was considered to be more common, but a study has since reported that just 14 out of 87 styrene-utilizing strains encode this pathway, indicating that direct ring cleavage, or a modified version of this pathway, is more widespread than initially thought [95]. Direct ring cleavage includes an initial dihydroxylation of the aromatic ring to form styrene cis-glycol, followed by actual ring cleavage (Fig. 5) and further degradation via the ortho or meta cleavage pathway of aromatic compounds, to derive central intermediates such as pyruvate, acrylic acid and acetaldehyde [93, 94, 96, 97]. Key enzymes in direct ring cleavage include styrene dioxygenase, which catalyses the first step in this pathway but currently lacks gene sequences, and glycol dehydrogenase (GDH) mediating the subsequent conversion to vinylcatechol (Fig. 5). We detected GDH homologues in nearly all of our samples (Table S13), indicating that this dehydrogenase is involved in many aspects of aromatic compound metabolism, and probably not restricted to styrene degradation.
In the second pathway, vinyl side chain oxygenation, styrene is degraded to styrene oxide via monooxygenases and eventually to phenylacetic acid (Fig. 5), leaving the aromatic ring intact [92, 98–100]. We detected genes for a complete styrene monooxygenase styA/styB (Fig. 5) solely in the PS faeces group (Table S13), probably because faeces represent an aerobic environment. To assess the evolutionary history of styrene monooxygenase, we inferred the phylogeny of styA genes and found that, while the overall gene phylogeny aligns with taxonomic classifications, there is evidence for considerable horizontal gene transfer (Fig. S9). StyA genes recovered from the superworm faeces were grouped with experimentally verified homologues, e.g. from Pseudomonas fluorescens [98], suggesting that they indeed encode functional styrene-degrading enzymes. Furthermore, homologues of styB, without the presence of styA, were detected in the bran and starvation group, indicating that these gene homologues encode enzymes functioning in pathways unrelated to styrene degradation. Also, styD genes, encoding phenylacetaldehyde dehydogenase (Fig. 5), were present in high numbers in all feeding trial groups (Table S13), confirming previous findings that styD is strongly homologous to a range of prokaryotic aldehyde dehydrogenases [98]. Distant homologues of styE genes, encoding an outer membrane protein reported to be involved in styrene transport in Pseudomonas putida [101], were detected in PS faeces and bran assemblies (Table S14). Phylogenetic inferences clustered the superworm microbiome styE homologues with genes for toluene catabolism membrane proteins [102] in a sister clade to a clade containing the originally described styE gene (Fig. S10). Therefore, styE might be involved in styrene transport in multiple genera, but functional validations are required to determine the role of the styE homologues recovered from the superworm gut.
Anaerobic transformation of styrene has been observed in microbial consortia isolated from sewage sludge, and degradation routes via phenylacetaldehyde and phenylacetate, similar to side chain oxidation, or via ethylphenol have been proposed [93, 103]. However, the involved enzymes remain to be characterized, and hence we could not assess their presence in the superworm gut microbiome.
Linking phylogeny and function
To associate the inferred metabolic capabilities of the superworm gut microbiome with the taxa observed in our community profiles, we binned MAGs from assemblies of all samples. We recovered 34 medium- to high-quality MAGs with an average estimated completeness of 90.0% (±9.7%) and a mean estimated contamination of 1.3% (±0.9%) (Table 1). In addition, we also included one low-quality draft MAG (48.8% estimated completeness) harbouring potential styrene degradation genes (Table 1). Taxonomic classification assigned the MAGs to 21 genera in four phyla: Proteobacteria , Firmicutes , Actinobacteria and Bacteroidota (Table 1, Fig. S11). MAGs assigned to the phylum Bacteroidota , orders Flavobacteriales and Shingobacteriales, and to the phylum Actinobacteria were solely recovered from PS faeces (Fig. S11), which was expected given the higher relative abundances of these lineages in the PS faeces community profile (Fig. 2).
Table 1.
MAG statistics
MAGs recovered from the polystyrene (ps), bran (bran) and starvation control (nf) groups, as well as from the polystyrene faeces (feces_ps) and the bran faeces (feces_b). Taxonomic classifications were carried out with GTDB-Tk (R202) and estimated completeness, estimated contamination and strain heterogeneity were assessed with CheckM
|
MAG ID |
Taxonomy |
Comp |
Cont |
Strain |
Q |
|---|---|---|---|---|---|
|
ps_bin.7 |
Lactococcus garvieae _A ( Firmicutes ; Bacilli ) |
86.11 |
0 |
0 |
86.11 |
|
ps_bin.4 |
Pseudomonas aeruginosa ( Proteobacteria ; Gammaproteobacteria ) |
48.8 |
0.56 |
0 |
46 |
|
ps_bin.3 |
Enterococcus _C sp009933135 ( Firmicutes ; Bacilli ) |
85.26 |
1.62 |
28.57 |
77.16 |
|
nf_max107.003 |
LSJC7 sp005671395 ( Proteobacteria ; Gammaproteobacteria ) |
99.03 |
0.74 |
0 |
95.33 |
|
nf_bin.6 |
Klebsiella _A oxytoca ( Proteobacteria ; Gammaproteobacteria ) |
98.2 |
0.19 |
50 |
97.25 |
|
nf_bin.5 |
Enterococcus _C sp009933135 ( Firmicutes ; Bacilli ) |
74.94 |
2.69 |
50 |
61.49 |
|
nf_bin.1 |
Enterococcus _D ( Firmicutes ; Bacilli ) |
96.62 |
2.51 |
7.69 |
84.07 |
|
feces_ps_gm2_bin_9 |
Klebsiella aerogenes ( Proteobacteria ; Gammaproteobacteria ) |
77.78 |
3.11 |
50 |
62.23 |
|
feces_ps_gm2_bin_7 |
Pseudomonas_E qingdaonensis ( Proteobacteria ; Gammaproteobacteria ) |
97.88 |
0.88 |
28.57 |
93.48 |
|
feces_ps_gm2_bin_6 |
Pseudomonas _E sp001320525 ( Proteobacteria ; Gammaproteobacteria ) |
93.56 |
1.31 |
25 |
87.01 |
|
feces_ps_bin.9 |
90.13 |
1.76 |
0 |
81.33 |
|
|
feces_ps_bin.7 |
91.91 |
0 |
0 |
91.91 |
|
|
feces_ps_bin.6 |
Rhodococcus hoagie ( Actinobacteriota ; Actinomycetia ) |
91.51 |
2.95 |
3.23 |
76.76 |
|
feces_ps_bin.5 |
97.88 |
1.35 |
33.33 |
91.13 |
|
|
feces_ps_bin.3 |
Pseudomonas aeruginosa ( Proteobacteria ; Gammaproteobacteria ) |
85.28 |
1.38 |
11.11 |
78.38 |
|
feces_ps_bin.21 |
Lactococcus garvieae _A ( Firmicutes ; Bacilli ) |
98.09 |
0.2 |
50 |
97.09 |
|
feces_ps_bin.2 |
98.12 |
0.83 |
0 |
93.97 |
|
|
feces_ps_bin.19 |
Enterococcus _C sp009933135 ( Firmicutes ; Bacilli ) |
90.81 |
1.98 |
15.38 |
80.91 |
|
feces_ps_bin.16 |
Brevundimonas ( Proteobacteria ; Alphaprteobacteria) |
83.17 |
0.35 |
50 |
81.42 |
|
feces_ps_bin.15 |
89.53 |
1.94 |
25 |
79.83 |
|
|
feces_ps_bin.14 |
98.42 |
0.56 |
0 |
95.62 |
|
|
feces_ps_bin.12 |
95.4 |
0.46 |
0 |
93.1 |
|
|
feces_ps_bin.10 |
97.86 |
1.46 |
0 |
90.56 |
|
|
feces_ps_bin.1 |
Pediococcus ( Firmicutes ; Bacilli ) |
73.2 |
1.81 |
16.67 |
64.15 |
|
feces_b_bin.9 |
LSJC7 sp005671395 ( Proteobacteria ; Gammaproteobacteria ) |
78.28 |
1.82 |
27.78 |
69.18 |
|
feces_b_bin.8 |
Enterobacter oligotrophicus ( Proteobacteria ; Gammaproteobacteria ) |
62.79 |
0 |
0 |
62.79 |
|
feces_b_bin.5 |
98.16 |
0.15 |
0 |
97.41 |
|
|
feces_b_bin.14 |
Spiroplasma _A ( Firmicutes ; Bacilli ) |
93.96 |
2.01 |
25 |
83.91 |
|
feces_b_bin.12 |
Lactococcus lactis_A ( Firmicutes ; Bacilli ) |
98.3 |
0 |
0 |
98.3 |
|
feces_b_bin.10 |
Enterococcus_B faecalis_A ( Firmicutes ; Bacilli ) |
94.26 |
0.37 |
0 |
92.41 |
|
bran_bin.7 |
LSJC7 ( Proteobacteria ; Gammaproteobacteria ) |
75.68 |
0.58 |
0 |
72.78 |
|
bran_bin.3 |
Enterobacter oligotrophicus ( Proteobacteria ; Gammaproteobacteria ) |
94.86 |
1.48 |
47.37 |
87.46 |
|
bran_bin.2 |
LSJC7 sp005671395 ( Proteobacteria ; Gammaproteobacteria ) |
99.01 |
1.74 |
15 |
90.31 |
|
bran_bin.11 |
Spiroplasma_A ( Firmicutes ; Bacilli ) |
74.61 |
2.01 |
0 |
64.56 |
|
bran_bin.10 |
99.42 |
2.76 |
18.92 |
85.62 |
Estimated completeness (by CheckM)=Comp; estimated contamination (by CheckM)=Cont; estimated strain heterogeneity (by CheckM)=Strain. The genome quality (Q) is calculated as Comp −5 x Cont. The only low-quality MAG (ps_bin.4) included in our analysis is highlighted in italic. Detailed taxonomy strings for each MAG are provided in Table S18. MAG ID colour code: PS and PS faeces (red), starvation control group (green), bran and bran faeces (blue).
Screening MAGs for inferred functions associated with PS and styrene degradation revealed that genes encoding triacylglycerol lipase, an enzyme that could be involved in the breakdown of partially degraded PS (see ‘Detection of potential polystyrene and styrene degrading enzymes’), were only present in MAGs from three classes ( Gammaproteobacteria , Actinomycetia , Bacteroidia ) recovered from the PS group and the PS faeces (Table S15). Within the Gammaproteobacteria , all triacylglycerol lipase encoding MAGs were assigned to species in the family Pseudomonadaceae , including Pseudomonas aeruginosa and Pseudomonas_E qingdaonensis (Table S15). MAGs from these taxa also possessed genes for SH (Table S16), an enzyme associated with carbonyl group-targeted PS degradation [39], for homologues of styE (Table S14), involved in styrene transport, and for glycol dehydrogenase (Table S15), a key enzyme in direct ring cleavage (Fig. 5). Based on these results, we conclude that Pseudomonadaceae strains are promising targets for future studies investigating PS- and styrene-degrading enzymes and pathways. This conclusion is supported by reports of P. aeruginosa strains growing on expanded PS films without additional carbon sources [104], and of P. aeruginosa strains degrading PS–poly lactic acid composites [105]. Furthermore, P. aeruginosa strains have been shown to thrive on crude oil [106], indicating that this species is able to break down a range of hydrocarbons.
Other MAGs possessing lipase genes were assigned to the Bacteroidota genus Sphingobacterium and to the Actinobacteria genera Rhodococcus and Corynebacterium . MAGs from the latter genus also encode styA (Table S15), the styrene monooxygenase mediating the first step in the vinyl side chain degradation (Fig. 5). Several Corynebacterium strains, including one identified as C. pseudodiphtheriticum based on morphological and physiological properties, have been isolated on styrene, and the authors report that styrene monooxygenase (styA) and styrene isomerase (styC) are located in the membrane of Corynebacterium species [107]. Therefore, members of this genus should not only be considered as opportunistic pathogens (see ‘Microbial community composition’), but also be included in future investigations of microbial PS-degrading pathways.
Alternative microbial carbon sources during PS feeding trials
Styrofoam, i.e. the white EPA used as PS source in our study, is manufactured by adding an expanding agent, in most cases the alkane pentane (C5H12), and the flame retardant hexabromocyclododecane (HBCD) [108]. The EPA used in our study contained 4–7% pentane and up to 1% HBCD at the point of production, according to the manufacturer (see Material and Methods). Therefore, we investigated if these compounds could function as carbon or energy sources for the microbial superworm gut community.
The flame retardant HBCD is a persistent organic pollutant that has been detected in a diverse range of environments, wildlife and humans, and causes developmental neurotoxicity in animals [109]. Several bacterial strains, including Pseudomonas sp. HB01, P. aeruginosa HS9 and Rhodopseudomonas palustris , were reported to effectively degrade HBCD [110]. While most chemical compounds produced during bacterial HBCD degradation have been studied in detail [111, 112], only two enzymes, a dehydrogenase and a transferase, involved in catalysing these reactions have been identified in R. palustris [112]. We screened our data for these two genes and while we did not detect homologues of the gene encoding 2-haloacid dehydrogenase (K01560), we found genes for glutathione S-transferase (K00799) in all feeding trials and faeces samples (Table S9). Since both enzymes are thought to work together sequentially [112], we conclude that an R. palustris-like pathway of HBCD degradation might not be present in the superworm gut microbiome. The presence of glutathione S-transferase genes in all our samples further suggests that this glutathione transferase is widely distributed in prokaryotes and is involved in a variety of processes, as suggested previously [113]. However, further transcriptomics and proteomics analysis is required to test for microbial HBCD degradation capabilities and their implications on PS feeding trials of insect larvae.
The ability to degrade alkanes, which include the expanding agent pentane, has been reported for several Pseudomonas strains, and enzymes implicated in this process include alkane monooxygenase (alkB), cytochrome P450 monooxygenase and flavin-binding monooxygenase (almA) [114, 115, 116]. Alkane degradation pathways usually start with the oxidation of a terminal methyl group yielding a primary alcohol, which is further oxidized to an aldehyde, converted into a fatty acid and finally oxidized through beta-oxidation to render CO2 [115]. We did not detect any of the involved monooxygenase genes in our samples, except for alkB genes in the PS group and the PS faeces (Table S17). The absence of a complete pentane degradation pathway in the superworm gut could be due to the limited oxygen availability in the intestine, as all aforementioned alkane degradation pathways rely on oxygen for substrate activation and as the terminal electron acceptor [115]. Anaerobic microbial alkane degradation pathways exist and have been reported to involve novel hydrocarbon activation mechanisms [117], although the involved enzymes remained unexplored. To our knowledge, the only anaerobic activation of alkanes studied on a transcriptional level involved Desulfatibacillum alkenivorans AK-01 [118]. The proposed key enzyme is an alkylsuccinate synthase (ass), which initiates alkane degradation via the addition of a fumarate molecule. Ass homologues were detected in the bran, PS and starvation feeding trials, and in the bran and PS faecal samples (Table S17). Based on our findings, it is possible that members of the superworm gut microbiome use pentane as a carbon source or for energy conservation. However, pentane availability in styrofoam is probably limited. Pentane diffuses rapidly after manufacture to less than 1% (wt) content over a period of about 2–3 weeks, and the remaining pentane continues to diffuse to insignificant levels within several weeks after production [26]. Regardless, further studies are needed to explore the abilities of microbial growth on alkenes, such as pentane, in the superworm gut.
Conclusion and outlook
Our study provides the first metagenomic analysis of the gut microbiome from the superworm (Zophobas morio), comparing microbial communities of worms reared on PS, on regular bran feed and under starvation conditions. The diet consisting solely of PS had considerable influences on the microbiome beyond the detection of PS degradation capabilities. It was characterized by a loss of microbial diversity, the detection of opportunistic pathogens indicative of dysbiosis, and an enrichment of inferred functions for stress response. These modifications of the gut microbiome, in combination with the minimal weight gain of the superworms reared on PS, paints a picture of survival under poor health conditions for the insect host. The presence of inferred PS- and styrene-degrading functions supports previous studies reporting PS-degrading abilities of the superworm microbiome. However, we cannot rule out that the microbes also metabolized other styrofoam components, such as the flame retardant or the remains of the expanding agent.
Our results support previous suggestions that superworms can help to reduce PS waste [39]. However, the minimal weight gain of the larvae on a PS diet will probably hamper their use in the PS recycling process. In particular, downstream applications such as biodiesel production from superworm fatty esters, an approach that has been proven feasible using superworms raised on regular feed [119], might not be achievable. Diet diversification, for example by supplementing styrofoam with food waste, could help to counteract the dietary deficits of the unbalanced PS feed and might increase gut microbiome health and subsequently host weight gain. Such an approach could also contribute towards waste valorization of agricultural and industrial by-products by insects [120]. Alternatively, isolating PS-degrading microbes, and characterizing their enzymes involved in PS degradation pathways, followed by enzyme engineering and large-scale production, are possible options to utilize the superworm microbiome.
In summary, our metagenomic exploration of the superworm gut microbiome provided the first insights into how their microbial community responds to a PS diet. However, comparable to opening a can of worms, our results also raised many new questions. Which members of the microbial community are active, and which genes are transcribed during a PS diet compared to regular feed? What are the complete pathways of PS and styrene degradation used by the gut microbes? Can some bacteria conserve energy by using styrofoam components other than PS? Employing transcriptomics to quantify gene expression levels [121], and using click chemistry to visualize in situ changes of translational activities [122] will contribute to answering these questions. However, bringing the majority of superworm gut bacteria into culture remains the ultimate goal in our endeavour to characterize microbial PS degradation in this and other insect microbiota.
Supplementary Data
Funding information
This work was primarily funded by The University of Queensland (UQ)/Australian Centre for Ecogenomics (ACE) strategic funding. C.R. was supported by an Australian Research Council (ARC) Future Fellowship (FT170100213). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Acknowledgements
We thank Phil Hugenholtz and Gene Tyson for their help in obtaining financial support for the project leading to this publication, and the ACE team (http://ecogenomic.org) for stimulating discussions.
Author contributions
Conceptualization by C.R.; Investigation by J.S. and C.R.; Formal Analysis by J.S., A.P. and C.R.; Data Curation by J.S.; Writing – Original Draft Preparation by J.S. and A.P.; Writing – Review and Editing by S.A. and C.R.; Visualization by J.S., A.P. and C.R.; Supervision by C.R.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: CAZy, carbohydrate active enzyme; EPA, expanded polystyrene foam; HBCD, hexabromocyclododecane; MAG, metagenome-assembled genome; PCA, principal component analysis; PE, polyethylene; PET, polyethylene terephthalate; PS, polystyrene; SH, serine hydrolase; SSU, small subunit.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Eleven supplementary figures and 18 supplementary tables are available with the online version of this article.
References
- 1.Thompson RC, Swan SH, Moore CJ, vom Saal FS. Our plastic age. Phil Trans R Soc B. 2009;364:1973–1976. doi: 10.1098/rstb.2009.0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Plastics Europe Plastics—the Facts 2019. Brussels, Belgium: 2019. An analysis of European plastics production, demand and waste data. [Google Scholar]
- 3.Barnes DKA, Galgani F, Thompson RC, Barlaz M. Accumulation and fragmentation of plastic debris in global environments. Phil Trans R Soc B. 2009;364:1985–1998. doi: 10.1098/rstb.2008.0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geyer R, Jambeck JR, Law KL. Production, use, and fate of all plastics ever made. Sci Adv. 2017;3:e1700782. doi: 10.1126/sciadv.1700782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plastics Europe Plastics - the Facts 2016. Brussels, Belgium: 2016. An analysis of European plastics production, demand and waste data. [Google Scholar]
- 6.Ho BT, Roberts TK, Lucas S. An overview on biodegradation of polystyrene and modified polystyrene: the microbial approach. Critical Reviews in Biotechnology. 2017;38:308–320. doi: 10.1080/07388551.2017.1355293. [DOI] [PubMed] [Google Scholar]
- 7.Kaplan DL, Hartenstein R, Sutter J. Biodegradation of polystyrene, poly(metnyl methacrylate), and phenol formaldehyde. Appl Environ Microbiol. 1979;38:551–553. doi: 10.1128/aem.38.3.551-553.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Otake Y, Kobayashi T, Asabe H, Murakami N, Ono K. Biodegradation of low-density polyethylene, polystyrene, polyvinyl chloride, and urea formaldehyde resin buried under soil for over 32 years. J Appl Polym Sci. 1995;56:1789–1796. doi: 10.1002/app.1995.070561309. [DOI] [Google Scholar]
- 9.Rochman CM, Tahir A, Williams SL, Baxa DV, Lam R, et al. Anthropogenic debris in seafood: Plastic debris and fibers from textiles in fish and bivalves sold for human consumption. Sci Rep. 2015;5:1–10. doi: 10.1038/srep14340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Albertsson A-C, Karlsson S. The influence of biotic and abiotic environments on the degradation of polyethylene. Progress in Polymer Science. 1990;15:177–192. doi: 10.1016/0079-6700(90)90027-X. [DOI] [Google Scholar]
- 11.O’Brine T, Thompson RC. Degradation of plastic carrier bags in the marine environment. Marine Pollution Bulletin. 2010;60:2279–2283. doi: 10.1016/j.marpolbul.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 12.Singh B, Sharma N. Mechanistic implications of plastic degradation. Polymer Degradation and Stability. 2008;93:561–584. doi: 10.1016/j.polymdegradstab.2007.11.008. [DOI] [Google Scholar]
- 13.Krueger MC, Seiwert B, Prager A, Zhang S, Abel B, et al. Degradation of polystyrene and selected analogues by biological Fenton chemistry approaches: Opportunities and limitations. Chemosphere. 2017;173:520–528. doi: 10.1016/j.chemosphere.2017.01.089. [DOI] [PubMed] [Google Scholar]
- 14.Gewert B, Plassmann MM, MacLeod M. Pathways for degradation of plastic polymers floating in the marine environment. Environ Sci Process Impacts. 2015;17:1513–1521. doi: 10.1039/c5em00207a. [DOI] [PubMed] [Google Scholar]
- 15.Wei R, Zimmermann W. Microbial enzymes for the recycling of recalcitrant petroleum‐based plastics: how far are we? Microb Biotechnol. 2017;10:1308–1322. doi: 10.1111/1751-7915.12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sielicki M, Focht DD, Martin JP. Microbial degradation of [ 14 C]polystyrene and 1,3-diphenylbutane. Can J Microbiol. 1978;24:798–803. doi: 10.1139/m78-134. [DOI] [PubMed] [Google Scholar]
- 17.Syranidou E, Karkanorachaki K, Amorotti F, Franchini M, Repouskou E, et al. Biodegradation of weathered polystyrene films in seawater microcosms. Sci Rep. 2017;7:17991. doi: 10.1038/s41598-017-18366-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang J, Yang Y, Wu W-M, Zhao J, Jiang L. Evidence of polyethylene biodegradation by bacterial strains from the guts of plastic-eating waxworms. Environ Sci Technol. 2014;48:13776–13784. doi: 10.1021/es504038a. [DOI] [PubMed] [Google Scholar]
- 19.Bombelli P, Howe CJ, Bertocchini F. Polyethylene bio-degradation by caterpillars of the wax moth Galleria mellonella . Current Biology. 2017;27:R292–R293. doi: 10.1016/j.cub.2017.02.060. [DOI] [PubMed] [Google Scholar]
- 20.Weber C, Pusch S, Opatz T. Polyethylene bio-degradation by caterpillars? Current Biology. 2017;27:R744–R745. doi: 10.1016/j.cub.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 21.Yang Y, Yang J, Wu W-M, Zhao J, Song Y, et al. Biodegradation and mineralization of polystyrene by plastic-eating mealworms: part 2. role of gut microorganisms. Environ Sci Technol. 2015;49:12087–12093. doi: 10.1021/acs.est.5b02663. [DOI] [PubMed] [Google Scholar]
- 22.Yang Y, Yang J, Wu W-M, Zhao J, Song Y, et al. Biodegradation and mineralization of polystyrene by plastic-eating mealworms: part 1. chemical and physical characterization and isotopic tests. Environ Sci Technol. 2015;49:12080–12086. doi: 10.1021/acs.est.5b02661. [DOI] [PubMed] [Google Scholar]
- 23.Brandon AM, Gao S-H, Tian R, Ning D, Yang S-S, et al. Biodegradation of Polyethylene and Plastic Mixtures in Mealworms (Larvae of Tenebrio molitor) and Effects on the Gut Microbiome. Environ Sci Technol. 2018;52:6526–6533. doi: 10.1021/acs.est.8b02301. [DOI] [PubMed] [Google Scholar]
- 24.Miao S-J, Zhang Y-L. Feeding and degradation effect on plastic of Zophobas morio . J Environ Entomol. 2010;X174 [Google Scholar]
- 25.Yang Y, Wang J, Xia M. Biodegradation and mineralization of polystyrene by plastic-eating superworms Zophobas atratus . Sci Total Environ. 2020;708:135233. doi: 10.1016/j.scitotenv.2019.135233. [DOI] [PubMed] [Google Scholar]
- 26.Simpson A, Rattigan I, Kalavsky E, Parr G. Thermal conductivity and conditioning of grey expanded polystyrene foams. Cellular Polymers. 2020;39:238–262. doi: 10.1177/0262489320934263. [DOI] [Google Scholar]
- 27.Maciel-Vergara G, Jensen AB, Eilenberg J. Cannibalism as a possible entry route for opportunistic pathogenic bacteria to insect hosts, exemplified by Pseudomonas aeruginosa, a pathogen of the giant mealworm Zophobas morio . Insects. 2018;9:88. doi: 10.3390/insects9030088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rumbos CI, Athanassiou CG. The Superworm, Zophobas morio (Coleoptera:Tenebrionidae): A “Sleeping Giant” in Nutrient Sources. J Insect Sci. 2021;21:13. doi: 10.1093/jisesa/ieab014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boyd JA, Woodcroft BJ, Tyson GW. GraftM: a tool for scalable, phylogenetically informed classification of genes within metagenomes. Nucleic Acids Res. 2018;46:e59. doi: 10.1093/nar/gky174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minich JJ, Sanders JG, Amir A, Humphrey G, Gilbert JA, et al. Quantifying and understanding well-to-well contamination in microbiome research. mSystems. 2019;4 doi: 10.1128/mSystems.00186-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Epstein HE, Smith HA, Cantin NE, Mocellin VJL, Torda G, et al. Temporal variation in the microbiome of Acropora coral species does not reflect seasonality. Front Microbiol. 2019;10 doi: 10.3389/fmicb.2019.01775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee MD, Walworth NG, Sylvan JB, Edwards KJ, Orcutt BN. Microbial communities on seafloor basalts at Dorado Outcrop reflect level of alteration and highlight global lithic clades. Front Microbiol. 2015;6:1470. doi: 10.3389/fmicb.2015.01470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 34.Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2014;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 36.Aramaki T, Blanc-Mathieu R, Endo H, Ohkubo K, Kanehisa M, et al. KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics. 2020;36:2251–2252. doi: 10.1093/bioinformatics/btz859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim HR, Lee HM, Yu HC, Jeon E, Lee S, et al. Biodegradation of polystyrene by Pseudomonas sp. isolated from the gut of superworms (Larvae of Zophobas atratus) Environ Sci Technol. 2020;54:6987–6996. doi: 10.1021/acs.est.0c01495. [DOI] [PubMed] [Google Scholar]
- 40.Bikandi J, Millan RS, Rementeria A, Garaizar J. In silico analysis of complete bacterial genomes: PCR, AFLP-PCR and endonuclease restriction. Bioinformatics. 2004;20:798–799. doi: 10.1093/bioinformatics/btg491. [DOI] [PubMed] [Google Scholar]
- 41.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH, Hancock J. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics. 2019 doi: 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weaver DK, McFarlane JE. The effect of larval density on growth and development of Tenebrio molitor . Journal of Insect Physiology. 1990;36:531–536. doi: 10.1016/0022-1910(90)90105-O. [DOI] [Google Scholar]
- 47.Gillott C. In: Entomology. Gillott C, editor. Dordrecht: Springer Netherlands; 1995. Postembryonic development; pp. 595–623. [Google Scholar]
- 48.Heiman ML, Greenway FL. A healthy gastrointestinal microbiome is dependent on dietary diversity. Molecular Metabolism. 2016;5:317–320. doi: 10.1016/j.molmet.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krams IA, Kecko S, Jõers P, Trakimas G, Elferts D, et al. Microbiome symbionts and diet diversity incur costs on the immune system of insect larvae. J Exp Biol. 2017;220:4204–4212. doi: 10.1242/jeb.169227. [DOI] [PubMed] [Google Scholar]
- 50.Priya NG, Ojha A, Kajla MK, Raj A, Rajagopal R. Host plant induced variation in gut bacteria of Helicoverpa armigera. PLoS One. 2012;7:e30768. doi: 10.1371/journal.pone.0030768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mattila HR, Rios D, Walker-Sperling VE, Roeselers G, Newton ILG, et al. Characterization of the active microbiotas associated with honey bees reveals healthier and broader communities when colonies are genetically diverse. PLoS ONE. 2012;7:e32962. doi: 10.1371/journal.pone.0032962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jung J, Heo A, Park YW, Kim YJ, Koh H, et al. Gut microbiota of Tenebrio molitor and their response to environmental change. J Microbiol Biotechnol. 2014;24:888–897. doi: 10.4014/jmb.1405.05016. [DOI] [PubMed] [Google Scholar]
- 53.Wang Y, Zhang Y. Investigation of gut-associated bacteria in Tenebrio molitor (Coleoptera: Tenebrionidae) larvae using culture-dependent and DGGE Methods. Ann Entomol Soc Am. 2015;108:941–949. doi: 10.1093/aesa/sav079. [DOI] [Google Scholar]
- 54.Urbanek AK, Rybak J, Wróbel M, Leluk K, Mirończuk AM. A comprehensive assessment of microbiome diversity in Tenebrio molitor fed with polystyrene waste. Environmental Pollution. 2020;262:114281. doi: 10.1016/j.envpol.2020.114281. [DOI] [PubMed] [Google Scholar]
- 55.Jurkevitch E. Riding the Trojan horse: combating pest insects with their own symbionts. Microb Biotechnol. 2011;4:620–627. doi: 10.1111/j.1751-7915.2011.00249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dillon RJ, Dillon VM. The gut bacteria of insects: nonpathogenic interactions. Annu Rev Entomol. 2004;49:71–92. doi: 10.1146/annurev.ento.49.061802.123416. [DOI] [PubMed] [Google Scholar]
- 57.Gupta A, Nair S. Dynamics of insect-microbiome interaction influence host and microbial symbiont. Front Microbiol. 2020;11:1357. doi: 10.3389/fmicb.2020.01357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gurung K, Wertheim B, Falcao Salles J. The microbiome of pest insects: it is not just bacteria. Entomol Exp Appl. 2019;167:156–170. doi: 10.1111/eea.12768. [DOI] [Google Scholar]
- 59.Samson R, Legendre JB, Christen R, Saux MF-L, Achouak W, et al. Transfer of Pectobacterium chrysanthemi (Burkholder et al. 1953) Brenner et al. 1973 and Brenneria paradisiaca to the genus Dickeya gen. nov. as Dickeya chrysanthemi comb. nov. and Dickeya paradisiaca comb. nov. and delineation of four novel species, Dickeya dadantii sp. nov., Dickeya dianthicola sp. nov., Dickeya dieffenbachiae sp. nov. and Dickeya zeae sp. nov. Int J Syst Evol Microbiol. 2005;55:1415–1427. doi: 10.1099/ijs.0.02791-0. [DOI] [PubMed] [Google Scholar]
- 60.Costechareyre D, Balmand S, Condemine G, Rahbé Y, Yang C-H. Dickeya dadantii, a plant pathogenic bacterium producing cyt-like entomotoxins, causes septicemia in the pea aphid Acyrthosiphon pisum . PLoS ONE. 2012;7:e30702. doi: 10.1371/journal.pone.0030702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fusco V, Quero GM, Cho G-S, Kabisch J, Meske D, et al. The genus Weissella: taxonomy, ecology and biotechnological potential. Front Microbiol. 2015;6:155. doi: 10.3389/fmicb.2015.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shuttleworth LA, Khan MAM, Osborne T, Collins D, Srivastava M, et al. A walk on the wild side: gut bacteria fed to mass-reared larvae of Queensland fruit fly [Bactrocera tryoni (Froggatt)] influence development. BMC Biotechnol. 2019;19:95. doi: 10.1186/s12896-019-0579-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dubin K, Pamer EG, Britton RA, Cani PD. Enterococci and their interactions with the intestinal microbiome. Microbiol Spectr. 2017;5 doi: 10.1128/microbiolspec.BAD-0014-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lebreton F, Willems RJL, Gilmore MS. In: Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. Gilmore MS, Clewell DB, Ike Y, Shankar N, editors. Boston: Massachusetts Eye and Ear Infirmary; 2014. Enterococcus diversity, origins in nature, and gut colonization. [PubMed] [Google Scholar]
- 65.Neely AN, Maley MP. Survival of enterococci and staphylococci on hospital fabrics and plastic. J Clin Microbiol. 2000;38:724–726. doi: 10.1128/JCM.38.2.724-726.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Balish E, Warner T. Enterococcus faecalis induces inflammatory bowel disease in interleukin-10 knockout mice. The American Journal of Pathology. 2002;160:2253–2257. doi: 10.1016/S0002-9440(10)61172-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wallen ZD, Appah M, Dean MN, Sesler CL, Factor SA, et al. Characterizing dysbiosis of gut microbiome in PD: evidence for overabundance of opportunistic pathogens. NPJ Parkinsons Dis. 2020;6:11. doi: 10.1038/s41531-020-0112-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Singh L, Cariappa MP, Kaur M. Klebsiella oxytoca: An emerging pathogen? Medical Journal Armed Forces India. 2016;72:S59–S61. doi: 10.1016/j.mjafi.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chow V, Nong G, Preston JF. Structure, function, and regulation of the aldouronate utilization gene cluster from Paenibacillus sp. strain JDR-2. J Bacteriol. 2007;189:8863–8870. doi: 10.1128/JB.01141-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Casacuberta E, González J. The impact of transposable elements in environmental adaptation. Mol Ecol. 2013;22:1503–1517. doi: 10.1111/mec.12170. [DOI] [PubMed] [Google Scholar]
- 71.Li S-J, Hua Z-S, Huang L-N, Li J, Shi S-H, et al. Microbial communities evolve faster in extreme environments. Sci Rep. 2014;4:6205. doi: 10.1038/srep06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vollmer W, Joris B, Charlier P, Foster S. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev. 2008;32:259–286. doi: 10.1111/j.1574-6976.2007.00099.x. [DOI] [PubMed] [Google Scholar]
- 73.Figueiredo TA, Sobral RG, Ludovice AM, Almeida JMF de, Bui NK, et al. Identification of genetic determinants and enzymes involved with the amidation of glutamic acid residues in the peptidoglycan of Staphylococcus aureus . PLoS Pathog. 2012;8:e1002508. doi: 10.1371/journal.ppat.1002508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zuber P. Spx-RNA polymerase interaction and global transcriptional control during oxidative stress. J Bacteriol. 2004;186:1911–1918. doi: 10.1128/JB.186.7.1911-1918.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Johnson V, Barbehenn R. Oxygen levels in the gut lumens of herbivorous insects. J Insect Physiol. 2000;46:897–903. doi: 10.1016/s0022-1910(99)00196-1. [DOI] [PubMed] [Google Scholar]
- 76.Valdes AM, Walter J, Segal E, Spector TD. Role of the gut microbiota in nutrition and health. BMJ. 2018:k2179. doi: 10.1136/bmj.k2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tegtmeier D, Thompson CL, Schauer C, Brune A. Oxygen affects gut bacterial colonization and metabolic activities in a gnotobiotic cockroach model. Appl Environ Microbiol. 2016;82:1080–1089. doi: 10.1128/AEM.03130-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stevenson L, Phillips F, O’Sullivan K, Walton J. Wheat bran: its composition and benefits to health, a European perspective. Int J Food Sci Nutr. 2012;63:1001–1013. doi: 10.3109/09637486.2012.687366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, et al. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell. 2016;167:1339–1353. doi: 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Audouin L, Langlois V, Verdu J, de Bruijn JCM. Role of oxygen diffusion in polymer ageing: kinetic and mechanical aspects. Journal of Materials Science. 2004;29:569–583. doi: 10.1007/BF00445968. [DOI] [Google Scholar]
- 81.Fontanella S, Bonhomme S, Koutny M, Husarova L, Brusson J-M, et al. Comparison of the biodegradability of various polyethylene films containing pro-oxidant additives. Polymer Degradation and Stability. 2010;95:1011–1021. doi: 10.1016/j.polymdegradstab.2010.03.009. [DOI] [Google Scholar]
- 82.Koutny M, Lemaire J, Delort A-M. Biodegradation of polyethylene films with prooxidant additives. Chemosphere. 2006;64:1243–1252. doi: 10.1016/j.chemosphere.2005.12.060. [DOI] [PubMed] [Google Scholar]
- 83.Lewis JC, Coelho PS, Arnold FH. Enzymatic functionalization of carbon-hydrogen bonds. Chem Soc Rev. 2011;40:2003–2021. doi: 10.1039/c0cs00067a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ma S, Preims M, Piumi F, Kappel L, Seiboth B, et al. Molecular and catalytic properties of fungal extracellular cellobiose dehydrogenase produced in prokaryotic and eukaryotic expression systems. Microb Cell Fact. 2017;16:37. doi: 10.1186/s12934-017-0653-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Frank A, Eborall W, Hyde R, Hart S, Turkenburg JP, et al. Mutational analysis of phenolic acid decarboxylase from Bacillus subtilis (BsPAD), which converts bio-derived phenolic acids to styrene derivatives. Catal Sci Technol. 2012;2:1568. doi: 10.1039/c2cy20015e. [DOI] [Google Scholar]
- 86.Mohan AJ, Sekhar VC, Bhaskar T, Nampoothiri KM. Microbial assisted high impact polystyrene (HIPS) degradation. Bioresource Technology. 2016;213:204–207. doi: 10.1016/j.biortech.2016.03.021. [DOI] [PubMed] [Google Scholar]
- 87.Chen C-C, Han X, Ko T-P, Liu W, Guo R-T. Structural studies reveal the molecular mechanism of PETase. FEBS J. 2018;285:3717–3723. doi: 10.1111/febs.14612. [DOI] [PubMed] [Google Scholar]
- 88.Eberl A, Heumann S, Brückner T, Araujo R, Cavaco-Paulo A, et al. Enzymatic surface hydrolysis of poly(ethylene terephthalate) and bis(benzoyloxyethyl) terephthalate by lipase and cutinase in the presence of surface active molecules. Journal of Biotechnology. 2009;143:207–212. doi: 10.1016/j.jbiotec.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 89.Kawai F, Kawabata T, Oda M. Current knowledge on enzymatic PET degradation and its possible application to waste stream management and other fields. Appl Microbiol Biotechnol. 2019;103:4253–4268. doi: 10.1007/s00253-019-09717-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang W, Cao H, Xu L, Zhang H, Yan Y. A novel eurythermic and thermostale lipase LipM from Pseudomonas moraviensis M9 and its application in the partial hydrolysis of algal oil. BMC Biotechnol. 2015;15:94. doi: 10.1186/s12896-015-0214-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Warhurst AM, Fewson CA. Microbial metabolism and biotransformations of styrene. Journal of Applied Bacteriology. 1994;77:597–606. doi: 10.1111/j.1365-2672.1994.tb02807.x. [DOI] [PubMed] [Google Scholar]
- 92.Bestetti G, Di Gennaro P, Colmegna A, Ronco I, Galli E, et al. Characterization of styrene catabolic pathway in Pseudomonas fluorescens ST. International Biodeterioration & Biodegradation. 2004;54:183–187. doi: 10.1016/j.ibiod.2004.06.005. [DOI] [Google Scholar]
- 93.Tischler D. In: Microbial Styrene Degradation, SpringerBriefs in Microbiology. Tischler D, editor. Cham: Springer International Publishing; 2015. Pathways for the degradation of styrene; pp. 7–22. [Google Scholar]
- 94.Warhurst AM, Clarke KF, Hill RA, Holt RA, Fewson CA. Metabolism of styrene by Rhodococcus rhodochrous NCIMB 13259. Appl Environ Microbiol. 1994;60:1137–1145. doi: 10.1128/aem.60.4.1137-1145.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Oelschlägel M, Zimmerling J, Schlömann M, Tischler D. Styrene oxide isomerase of Sphingopyxis sp. Kp5.2. Microbiology (Reading) 2014;160:2481–2491. doi: 10.1099/mic.0.080259-0. [DOI] [PubMed] [Google Scholar]
- 96.Patrauchan MA, Florizone C, Eapen S, Gómez-Gil L, Sethuraman B, et al. Roles of ring-hydroxylating dioxygenases in styrene and benzene catabolism in Rhodococcus jostii RHA1. J Bacteriol. 2008;190:37–47. doi: 10.1128/JB.01122-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tischler D, Kaschabek SR. In: Microbial Degradation of Xenobiotics, Environmental Science and Engineering. Singh SN, editor. Berlin, Heidelberg: Springer; 2012. Microbial styrene degradation: from basics to biotechnology; pp. 67–99. [Google Scholar]
- 98.Beltrametti F, Marconi AM, Bestetti G, Colombo C, Galli E, et al. Sequencing and functional analysis of styrene catabolism genes from Pseudomonas fluorescens ST. Appl Environ Microbiol. 1997;63:2232–2239. doi: 10.1128/aem.63.6.2232-2239.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Itch N, Hayashi K, Okada K, Ito T, Mizuguchi N. Characterization of styrene oxide isomerase, a key enzyme of styrene and styrene oxide metabolism in Corynehacterium sp. Biosci Biotechnol Biochem. 1997;61:2058–2062. doi: 10.1271/bbb.61.2058. [DOI] [PubMed] [Google Scholar]
- 100.O’Connor K, Buckley CM, Hartmans S, Dobson AD. Possible regulatory role for nonaromatic carbon sources in styrene degradation by Pseudomonas putida CA-3. Appl Environ Microbiol. 1995;61:544–548. doi: 10.1128/aem.61.2.544-548.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mooney A, O’Leary ND, Dobson ADW. Cloning and functional characterization of the styE Gene, involved in styrene transport in Pseudomonas putida CA-3. Appl Environ Microbiol. 2006;72:1302–1309. doi: 10.1128/AEM.72.2.1302-1309.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kahng H-Y, Byrne AM, Olsen RH, Kukor JJ. Characterization and role of tbuX in utilization of toluene by Ralstonia pickettii PKO1. J Bacteriol. 2000;182:1232–1242. doi: 10.1128/JB.182.5.1232-1242.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Grbić-Galić D, Churchman-Eisel N, Mraković I. Microbial transformation of styrene by anaerobic consortia. J Appl Bacteriol. 1990;69:247–260. doi: 10.1111/j.1365-2672.1990.tb01516.x. [DOI] [PubMed] [Google Scholar]
- 104.Atiq N, Ahmed S, Ali MI, Saadia leeb, Ahmad B, et al. Isolation and identification of polystyrene biodegrading bacteria from soil. Afr J Microbiol Res. 2010;4:1537–1541. [Google Scholar]
- 105.Shimpi N, Borane M, Mishra S, Kadam M. Biodegradation of polystyrene (PS)-poly(lactic acid) (PLA) nanocomposites using Pseudomonas aeruginosa . Macromol Res. 2012;20:181–187. doi: 10.1007/s13233-012-0026-1. [DOI] [Google Scholar]
- 106.Talaiekhozani A, Jafarzadeh N, Fulazzaky MA, Talaie MR, Beheshti M. Kinetics of substrate utilization and bacterial growth of crude oil degraded by Pseudomonas aeruginosa . J Environ Health Sci Eng. 2015;13:64. doi: 10.1186/s40201-015-0221-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Itoh N, Yoshida K, Okada K. Isolation and identification of styrene-degrading Corynebacterium strains, and their styrene metabolism. Biosci Biotechnol Biochem. 1996;60:1826–1830. doi: 10.1271/bbb.60.1826. [DOI] [PubMed] [Google Scholar]
- 108.Law RJ, Kohler M, Heeb NV, Gerecke AC, Schmid P, et al. Hexabromocyclododecane challenges scientists and regulators. Environ Sci Technol. 2005;39:281A–287A. doi: 10.1021/es053302f. [DOI] [PubMed] [Google Scholar]
- 109.Szabo DT. In: Encyclopedia of Toxicology. 3rd ed. Wexler P, editor. Oxford: Academic Press; 2014. Hexabromocyclododecane; pp. 864–868. [Google Scholar]
- 110.Li Y-J, Wang R, Lin C-Y, Chen S-H, Chuang C-H, et al. The degradation mechanisms of Rhodopseudomonas palustris toward hexabromocyclododecane by time-course transcriptome analysis. Chemical Engineering Journal. 2021;425:130489. doi: 10.1016/j.cej.2021.130489. [DOI] [Google Scholar]
- 111.Huang L, Wang W, Shah SB, Hu H, Xu P, et al. The HBCDs biodegradation using a Pseudomonas strain and its application in soil phytoremediation. Journal of Hazardous Materials. 2019;380:120833. doi: 10.1016/j.jhazmat.2019.120833. [DOI] [PubMed] [Google Scholar]
- 112.Wang R, Lin C-Y, Chen S-H, Lo K-J, Liu C-T, et al. Using high-throughput transcriptome sequencing to investigate the biotransformation mechanism of hexabromocyclododecane with Rhodopseudomonas palustris in water. Sci Total Environ. 2019;692:249–258. doi: 10.1016/j.scitotenv.2019.07.140. [DOI] [PubMed] [Google Scholar]
- 113.Allocati N, Federici L, Masulli M, Di Ilio C. Glutathione transferases in bacteria. FEBS J. 2009;276:58–75. doi: 10.1111/j.1742-4658.2008.06743.x. [DOI] [PubMed] [Google Scholar]
- 114.Liu H, Xu J, Liang R, Liu J. Characterization of the medium- and long-chain n-alkanes degrading Pseudomonas aeruginosa strain SJTD-1 and its alkane hydroxylase genes. PLoS One. 2014;9:e105506. doi: 10.1371/journal.pone.0105506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rojo F. Degradation of alkanes by bacteria. Environ Microbiol. 2009;11:2477–2490. doi: 10.1111/j.1462-2920.2009.01948.x. [DOI] [PubMed] [Google Scholar]
- 116.Vomberg A, Klinner U. Distribution of alkB genes within n-alkane-degrading bacteria. J Appl Microbiol. 2000;89:339–348. doi: 10.1046/j.1365-2672.2000.01121.x. [DOI] [PubMed] [Google Scholar]
- 117.Widdel F, Rabus R. Anaerobic biodegradation of saturated and aromatic hydrocarbons. Curr Opin Biotechnol. 2001;12:259–276. doi: 10.1016/s0958-1669(00)00209-3. [DOI] [PubMed] [Google Scholar]
- 118.Herath A, Wawrik B, Qin Y, Zhou J, Callaghan AV, et al. Transcriptional response of Desulfatibacillum alkenivorans AK-01 to growth on alkanes: insights from RT-qPCR and microarray analyses. FEMS Microbiology Ecology. 2016;92:fiw062. doi: 10.1093/femsec/fiw062. [DOI] [PubMed] [Google Scholar]
- 119.Leung D, Yang D, Li Z, Zhao Z, Chen J, et al. Biodiesel from Zophobas morio larva oil: process optimization and FAME characterization. Ind Eng Chem Res. 2012;51:1036–1040. doi: 10.1021/ie201403r. [DOI] [Google Scholar]
- 120.Derler H, Lienhard A, Berner S, Grasser M, Posch A, et al. Use them for what they are good at: mealworms in circular food systems. Insects. 2021;12:40. doi: 10.3390/insects12010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hawley AK, Nobu MK, Wright JJ, Durno WE, Morgan-Lang C, et al. Diverse Marinimicrobia bacteria may mediate coupled biogeochemical cycles along eco-thermodynamic gradients. Nat Commun. 2017;8:1507. doi: 10.1038/s41467-017-01376-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lindivat M, Larsen A, Hess-Erga OK, Bratbak G, Hoell IA. Bioorthogonal non-canonical amino acid tagging combined with flow cytometry for determination of activity in aquatic microorganisms. Front Microbiol. 2020;11:1929. doi: 10.3389/fmicb.2020.01929. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.