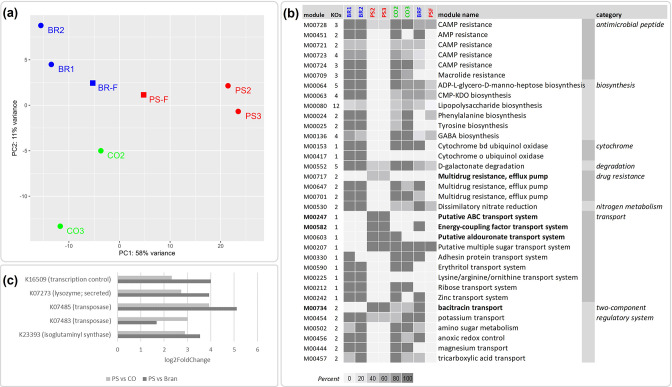
Fig. 4.
Functional profiles of superworm microbiomes. (a) PCA of genes assigned to bacterial KEGG Orthologs (KO identifiers), showing a clear separation of all three feeding trials, including bran (BR; blue dots), polystyrene (PS; red dots), starvation control group (CO; green dots), as well as polystyrene faeces (PSF; red squares) and bran faeces (BRF; blue squares). (b) KEGG modules enriched and depleted in the polystyrene (PS) group based on rarefied gene counts. Shown are the KEGG modules, the number of KOs per module and the module completeness (0–100 %) reported for the bran samples (BR1, BR2), polystyrene samples (PS2, PS3), and the starvation control samples (CO2, CO3). Modules present in the PS group but absent in the bran group are highlighted in bold. (c) Differential abundant genes, with KO identifiers, enriched in the PS group compared to the starvation control (CO) and bran group, respectively. Note that gene counts were rarefied to 8255 for the module comparison in (b).